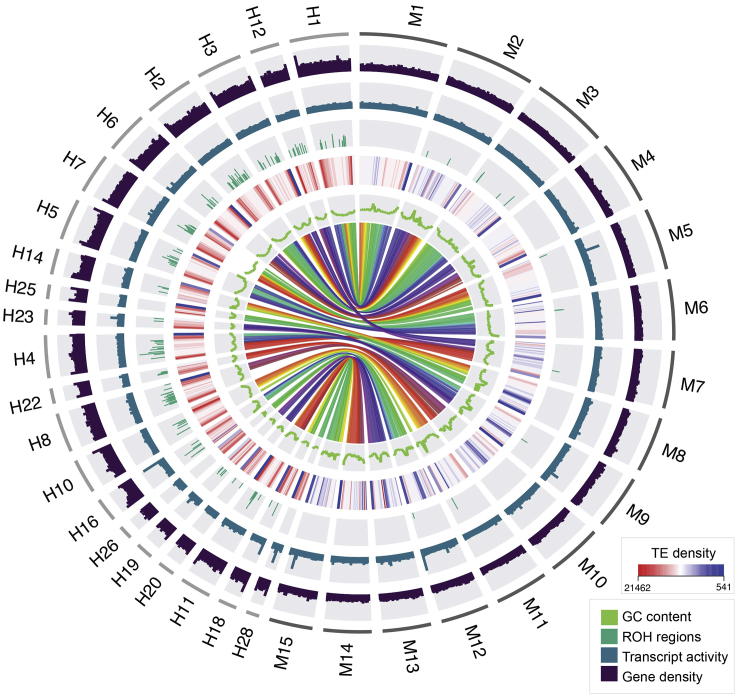
Figure 1.
Circos plot representation of genomic features in great hammerhead (H) and shortfin mako (M) for a subset of the largest scaffolds
Only a subset of scaffolds is depicted here in order to provide resolution of genome characteristics. The outside ring indicates different chromosomes; the 15 largest chromosomes are represented for shortfin mako, with chromosomes syntenic to those 15, represented for great hammerhead. Gene density is shown in the next, black colored ring, followed by transcript activity in blue (calculated by aligning RNA-seq to the reference genome and normalizing to read counts per million (RPM)); the green colored ring illustrates ROH>1Mbp in histogram bars, the height of which depicts the relative length of these ROH. TE density is plotted by a heatmap representation, GC content in the inside green line, and multicolored syntenic regions between great hammerhead and shortfin mako are illustrated in the middle.