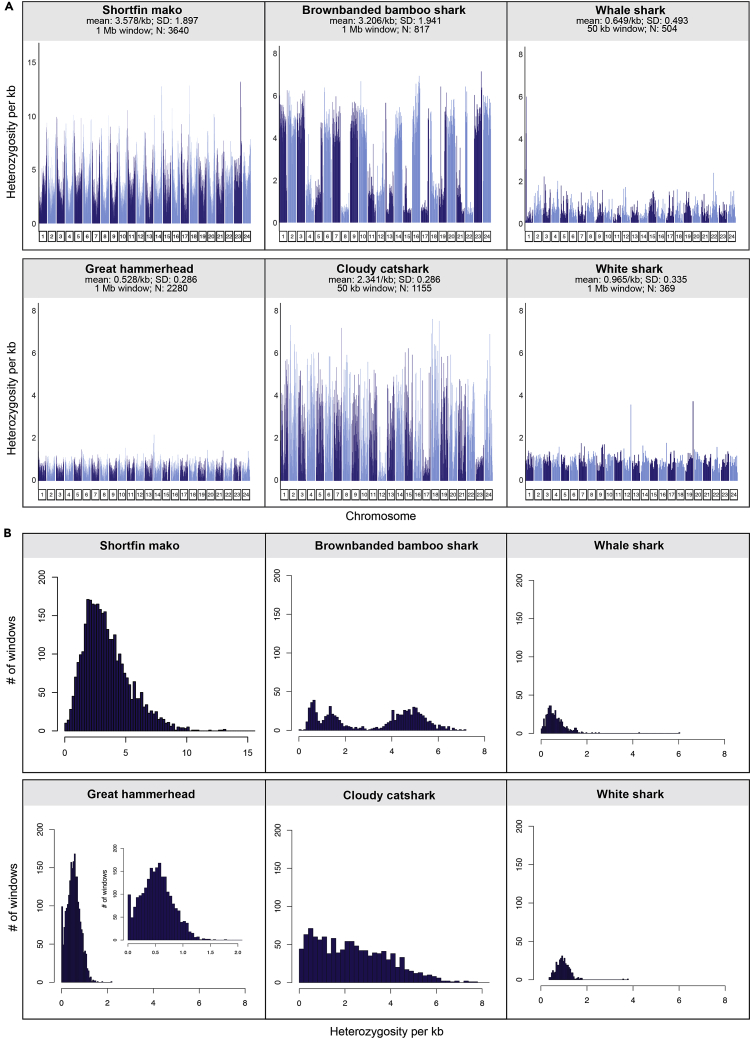
Figure 2.
Genome heterozygosity across the 24 largest scaffolds for several species of sharks
(A) Heterozygosity plotted for each of the scaffolds for each species, at non-overlapping windows of 1 Mbp for great hammerhead, shortfin mako, brownbanded bamboo shark, and white shark, and at 50 kbp windows for cloudy catshark and whale shark; shorter windows chosen for these latter two species because of their more fragmented draft genomes. Although the white shark is also a draft genome, there were nonetheless sufficient windows that could be sampled at 1 Mbp, to allow for accurate heterozygosity estimates. N refers to number of windows sampled. Complete genome-wide heterozygosity for all 40 and 41 pseudo-chromosomes of the great hammerhead and shortfin mako, respectively, appear in Figure S3.
(B) Histograms of per window heterozygosity derived from the set of 24 scaffolds for each species; the smaller distribution for whale shark and white shark reflects the smaller number of windows sampled for these two species.