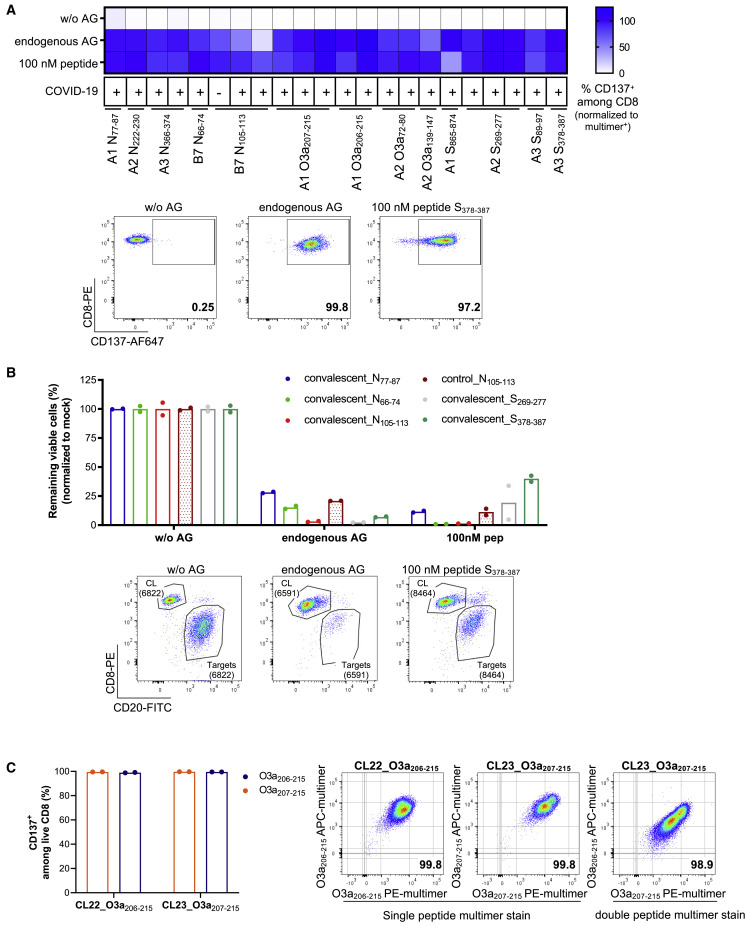
Figure 5.
Functional validation of CD8 epitopes
(A and B) Functional validation of immunogenic peptides by co-culture of multimer positive T cell lines derived from 11 convalescents and 1 pre-pandemic individual (indicated by COVID-19 status as + or –, respectively) for 20 h with mono-allelic B721.221 cells transduced to express the relevant protein (endogenous antigen [AG]), or not (without AG), or loaded with 100 nM peptide as a positive control (mean of technical duplicates or triplicates).
(A) Activation of T cells was determined as the percent of CD137+ cells among live CD8. The percent response is normalized to the percent multimer+ of live CD8 T cells in each T cell line. Representative flow plots are shown for the MS identified peptide S378–387.
(B) Target cell killing mediated by multimer+ T cell lines. Values are normalized to matching mock control and displayed as percent remaining viable cells. Representative flow plots for the MS identified peptide S378–387 show CD20+ B721.221 target cells and CD8+ multimer positive T cell line (CL) (numbers represent absolute counts).
(C) Cross-recognition of T cell lines sorted from one convalescent for reactivity to ORF3a-derived peptides O3a207–215 (FTSDYYQLY; 9-mer) or O3a206–215 (YFTSDYYQLY; 10-mer) to the other peptide. CL22 (sorted for reactivity to O3a206–215) and CL23 (sorted for reactivity to O3a207–215) recognize the 9- and 10-mer equally well (mean of technical duplicates). (Left) Activation marker expression (CD137) after 20 h co-culture with peptide-loaded (100 nM) mono-allelic B721.221 cells. (Right) Flow plots 1 and 2: dual-color multimer staining of CL22 and CL23 with multimers complexed with relevant peptide. Flow plot 3: multimer staining of CL23 with multimers complexed with 9-mer (x axis) and 10-mer (y axis).