Figure 3.
Phosphorylation dynamic analysis reveals potential CDK-driven events triggered by WEE1 inhibition
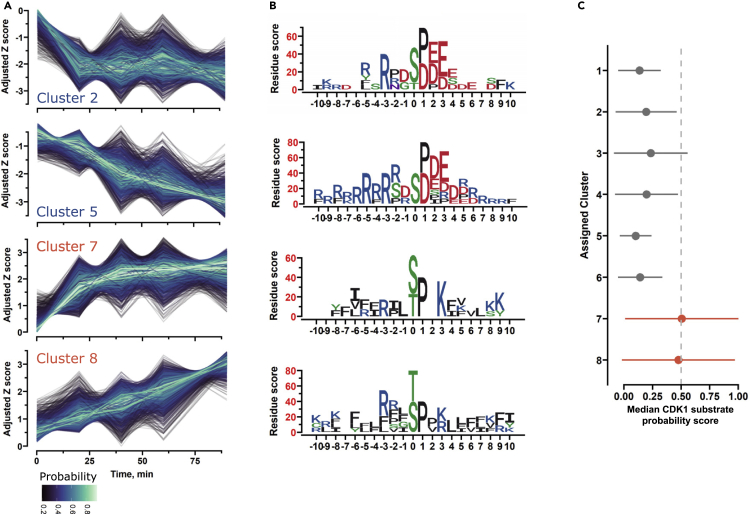
(A) Fuzzy-c-means (FCM) cluster profiles for select clusters, time is plotted on the x axis and adjusted Z scores on the y axis. Each line corresponds to the dynamics of a single phosphopeptide. Color intensity represents the probability of classification to the respective cluster. Z-scores are adjusted by adding or subtracting the minimal value.
(B) Phosphorylation sites sequence motifs for the clusters in A generated with a binomial probability model. x axis indicates the amino acid position from the phosphorylation site. Amino acid colors are based on chemical properties.
(C) Median CDK1 substrate probability scores are based on position-specific scoring matrixes (PSSM). Clusters 7 and 8 that may capture CDK-dependent events are indicated in red. The line indicates the median absolution deviation of the median CDK1 substrate probability score.