Figure 4.
Prediction of large number of CDK sites through time-resolved phosphoproteomic data in combination with previous knowledge databases
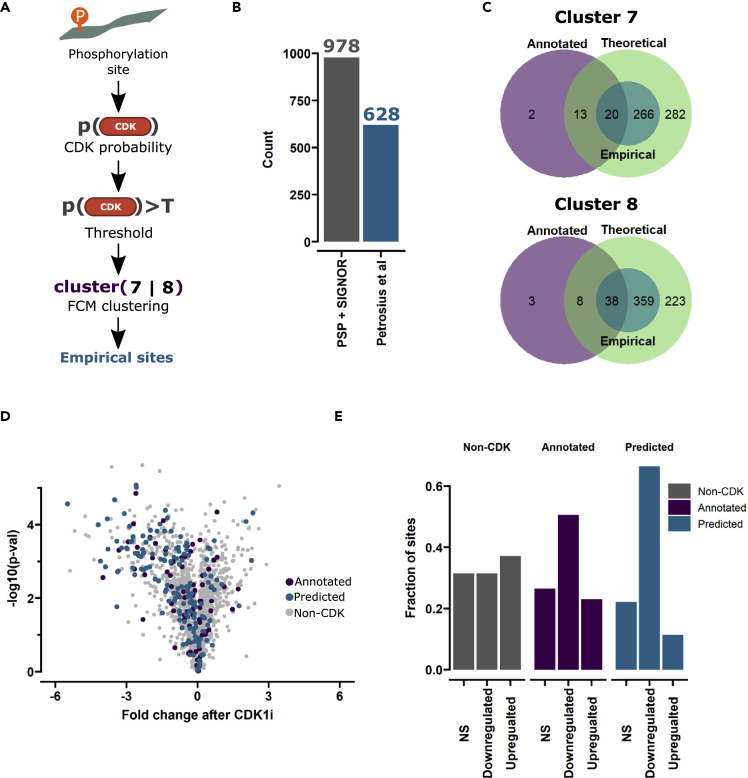
(A) Scheme for prediction of theoretical and empirical CDK sites. A probability for CDK1 or CDK2 is calculated for every quantified phosphorylation site, which is used to classify the sites into non-CDK and theoretical CDK sites. Experimental data are then taken into account, by further filtering the potential CDK list for FCM stratification and significant upregulation.
(B) Bar plot showing annotated CDK sites in PhosphoSitePlus (PSP) and SIGNOR databases and the number of predicted sites in this study (Petrosius et al.).
(C) Venn diagram showing the overlap between annotated (PSP and SIGNOR databases), theoretical (highest PSSM probability score for CDK1 or CDK2), and empirical CDK sites.
(D) Volcano plot, based on quantification data obtained from Petrone et al., 2016. The fold change is plotted on the x axis and the negative log10 of the p value on the y axis. Predicted and annotated CDK sites are marked with distinct colors.
(E) Bar plot showing fractions of non-CDK, annotated CDK, and predicted CDK sites that are not significantly altered (NS), upregulated, or downregulated after CDK1 inhibition.