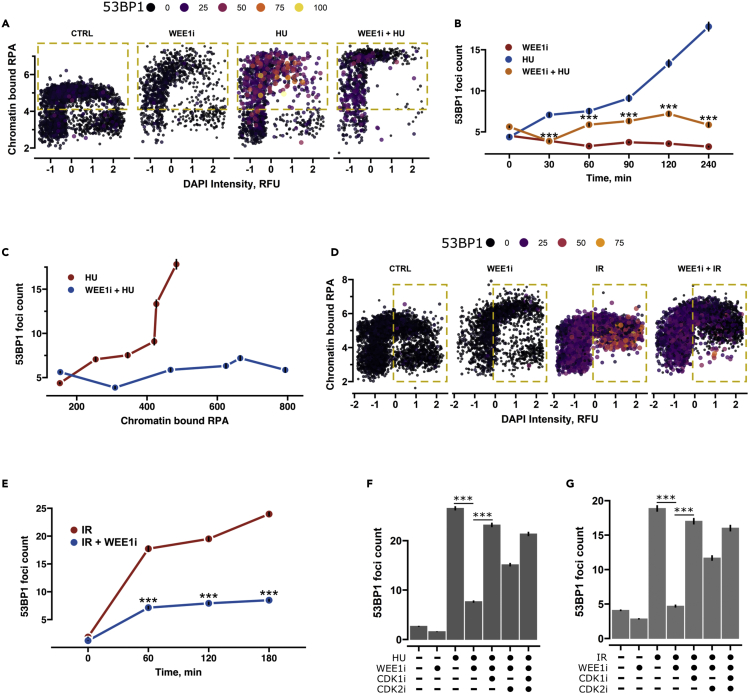
Figure 7.
WEE1 promotes the genotoxic stress-induced recruitment of 53BP1 to chromatin by limiting CDK activity
(A) Scatterplot showing cell cycle distribution based on DAPI and mean chromatin-bound RPA intensity. Color intensity represents 53BP1 foci count. Yellow dashed segment indicates the cell population used for summary statistic calculation. Cells were treated with 4 mM of hydroxyurea (HU) for 4 h, WEE1i (adavosertib) was used at 1 μM concentration. N = 2, n > 900.
(B) Dot plot showing 53BP1 foci mean values resolved by time with HU, WEE1i, or HU + WEE1i treatment. N = 2, n > 900, p-values were calculated with Kolmogorov-Smirnov test. ∗∗∗ indicates p-val <0.0001. HU and HU + WEE1i conditions are compared. Black bars indicate the SE.
(C) Plot showing the correlation between mean chromatin-bound RPA intensity and mean 53BP1 foci count. N = 2, n > 900, black bars indicate the SE.
(D and E) As in A and B, but cells were treated with 2 Gy of IR. The 3 h post IR time point is shown in D. N = 2, n > 2500.
(F and G) Bar plots showing mean 53BP1 foci values with addition of CDK1- (RO-3306 - 10 μM) and CDK2 (CDK2 inhibitor II - 5 μM)-specific inhibitors. Black bars indicate SE. N = 2, n > 500, p values were calculated with Kolmogorov-Smirnov test. ∗∗∗ indicates p-val <0.0001. All experiments were carried out at least in biological duplicate and one representative replicate is shown. N indicates the number of times the experiment has been replicated and n notes the minimal amount of cells quantified per condition.