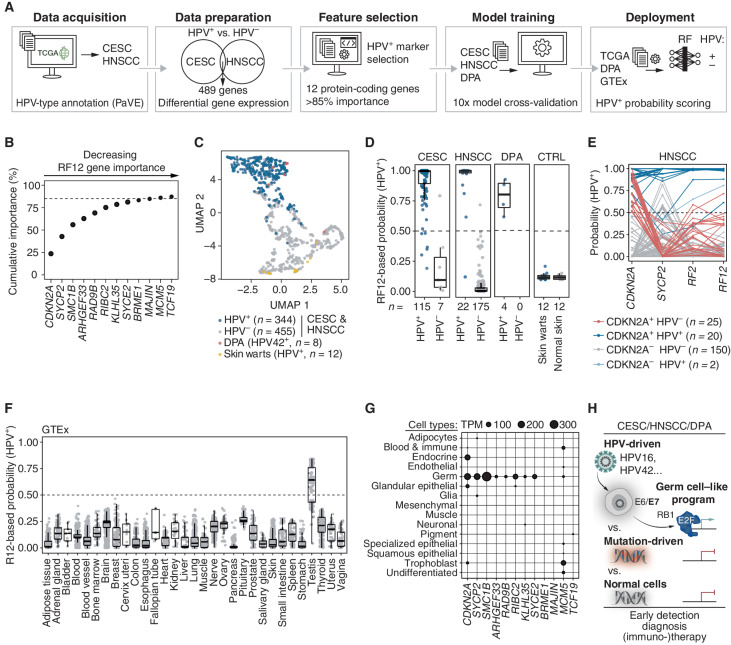
Figure 3.
Oncogenic HPVs induce a germ cell–like transcriptional program conserved throughout HPV-driven cancers. A, Overview of the random forest (RF) feature selection and machine learning procedure to identify a transcriptional fingerprint of HPV-driven oncogenesis. CESC and HNSCC expression data were annotated with HPV status (data acquisition), commonly differentially expressed genes between HPV+ and HPV− were identified (data preparation), and 12 protein-coding signature genes (RF12) were identified (feature selection). Four different machine learning models were trained with RF12 on a subset of DPA, CESC, and HNSCC samples (model training); the model with the best hyperparameters was evaluated using the withheld sample subset and used to classify Genotype-Tissue Expression (GTEx), skin warts, or normal skin expression data (deployment). B, Contribution of RF12 signature genes to cumulative feature importance (%) to discriminate between HPV+ and HPV− tumor samples. C, Uniform manifold approximation and projection (UMAP) dimensionality reduction of RF12 expression in DPA, CESC, HNSCC, and skin warts. D, HPV+ probability scores were calculated by RF12 for CESC, HNSCC, DPA, skin warts, and normal skin. For CESC, HNSCC, and DPA, the test set samples are displayed. E, HPV+ probability scores for HNSCC calculated for CDKN2A and SYCP2 alone, the combination of CDKN2A and SYCP2 (RF2), and RF12. F, HPV+ probability scores were calculated by RF12 for 31 normal tissues obtained from the GTEx database. G, Expression levels of RF12 genes in 15 cell types obtained from Human Protein Atlas. H, Schematic model of the germ cell–like program in HPV-driven cancer.