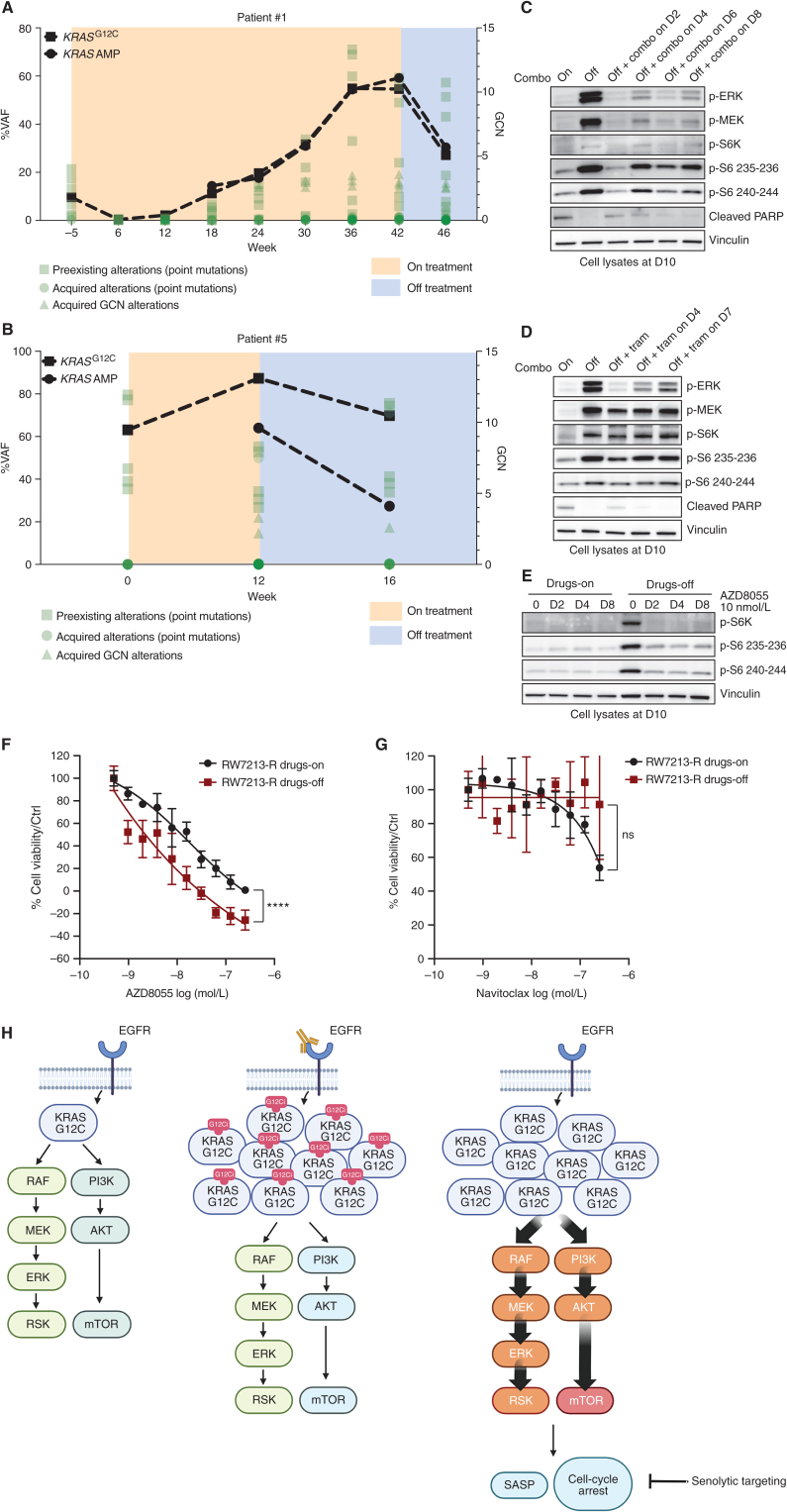
Figure 4.
Effect of treatment withdrawal in resistant colorectal cancers with amplified KRASG12C. A and B, Longitudinal analysis of ctDNA in colorectal cancer patients who held KRAS and EGFR inhibition for approximately 4 weeks after progression. KRASG12C ctDNA VAFs are marked with squares, and KRAS plasma copy numbers are marked with circles. All the other variants are reported in green. AMP, amplification; GCN, gene copy number. C, Western blot analyses of p-ERK, p-MEK, p-S6K, p-S6, and cleaved PARP expression upon drug withdrawal and rechallenge with 50 μg/mL cetuximab and 3 μmol/L sotorasib combination; vinculin is included as a loading control. D, Western blot analyses of p-ERK, p-MEK, p-S6K, p-S6, and cleaved PARP expression upon drug withdrawal and rechallenge with 10 nmol/L trametinib (tram); vinculin is included as a loading control. E, Western blot analyses of p-S6K and p-S6 upon drug withdrawal or in drug-containing medium after treatment with 10 nmol/L AZD8055; vinculin is included as a loading control.F, Short-term proliferation assay of RW7213 resistant cells (RW7213-R) in medium containing cetuximab–sotorasib (black) and in senescent conditions (dark red). Cells were seeded in the absence or presence of drugs for 4 days and then treated for 96 hours with increasing concentrations of AZD8055, and then ATP content was measured using CellTiter-Glo. Data represent the average and standard deviation of 3 biological replicates. ns (not significant) = P > 0.05; ****, P ≤ 0.0001. G, Short-term proliferation assay of RW7213-R in medium containing cetuximab–sotorasib (black) and in senescent conditions (dark red). Cells were seeded in the absence or presence of drugs for 4 days and then treated for 96 hours with increasing concentration of navitoclax, and then ATP content was measured using CellTiter-Glo. Data represent the average and standard deviation of 3 biological replicates. H, Proposed model: KRASG12C mutant signaling is maintained at a similar level in parental cells and in resistant cells in the presence of concomitant EGFR and KRASG12C blockade. Upon drug removal, KRASG12C-amplified signaling drives oncogene-induced senescence characterized by elevated mTOR activity, creating a new steady state that may be targeted by senolytic treatments.