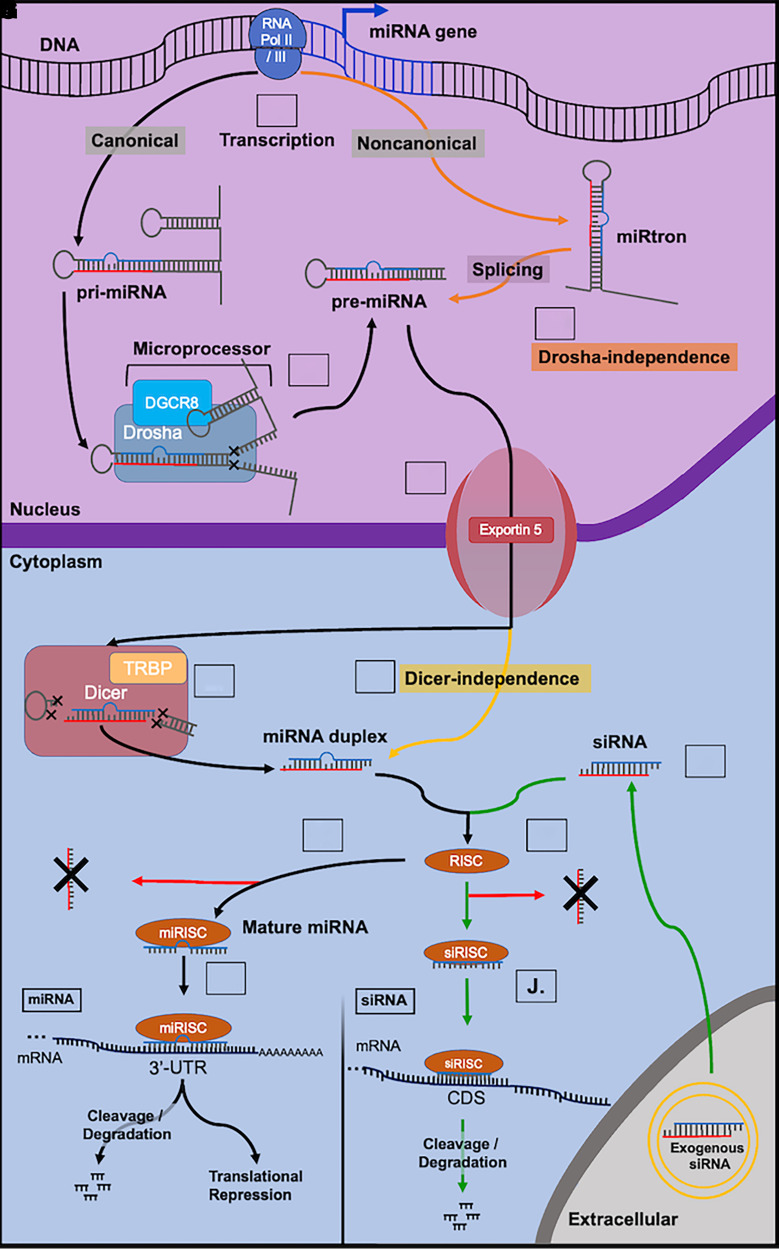
Fig. 1.
Overview of miRNA biogenesis and functions and siRNA mechanisms of action. (A) Intragenic or intergenic miRNA genes are transcribed by RNA polymerases II or III into primary miRNA (pri-miRNA; >1,000 nucleotides) transcripts in canonical pathway (black lines). (B) Pri-miRNAs are subjected to nuclear processing by the microprocessor Drosha-DGCR8 complex to release shorter precursor miRNAs (pre-miRNAs) (e.g., ∼65 nucleotides). (C) Noncanonical miRNA transcripts (e.g., mirtrons) are derived from the genome and subjected to RNA splicing to form pre-miRNAs independent on the microprocessor (orange lines). (D) Pre-miRNAs are exported into the cytoplasm via Exportin 5 transport complex. (E) In the cytoplasm, pre-miRNAs are processed by Dicer/TRBP complex to form miRNA duplexes (18–25 nucleotides). (F) Dicer-independent production of miRNA duplexes (yellow). (G) The guide strand (blue) of the miRNA duplex is selected and loaded into the RNA-induced silencing complex (RISC) to form the miRNA-RISC complex (miRISC) while the passenger strand (red) is degraded. (H) Functional miRNA binds to the 3′-untranslated region (3′UTR) of targeted mRNA to perform posttranscriptional gene regulation, either to accelerate mRNA cleavage or degradation or to repress translation. (I) Exogenous siRNAs can be introduced into cytoplasm through endocytosis or receptor-mediated uptake (green). (J) The antisense strand (blue) is selectively loaded into the RISC to form the siRNA-RISC complex (siRISC) while the sense strand (red) is degraded. (K) Functional siRNA typically acts on the protein coding sequence (CDS) of target transcript to cleave or initiate transcript degradation.