Figure 5. Targeting ZBTB2 homodimer formation as a rational strategy to inhibit the growth of functional p53‐deficient cancers.
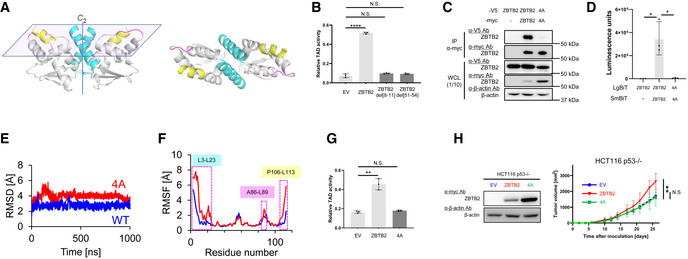
- A homology model of the N‐terminus of ZBTB2 based on the hMN crystal structure. L3‐23L (cyan), A86‐L89 (pink), and P106‐L113 (yellow). (Left) Side view. (Right) Top view.
- HCT116 p53−/− cells co‐transfected with pG5H1bLuc, pcDNA6/Gal4 DBD‐HIF‐1α TAD P564A, and pEF6/ZBTB2 (ZBTB2), pEF6/ZBTB2 del[8–11] (ZBTB2 del[8–11]), pEF6/ZBTB2 del[51–54] (ZBTB2 del[51–54]), or pEF6/myc‐His B (EV), as indicated, were cultured under < 0.1% oxygen conditions for 24 h and subjected to the luciferase assay.
- After transient transfection with or without the expression vectors for ZBTB2‐V5, ZBTB2 4A‐V5, ZBTB2‐myc, ZBTB2 4A‐myc, or their empty vector, as indicated, the myc‐fused proteins were immunoprecipitated using an anti‐myc antibody and co‐precipitated ZBTB2‐V5 or ZBTB2 4A‐V5 was detected (upper). One‐tenth of the whole‐cell lysate (WCL) was subjected to immunoblotting with the indicated antibodies (lower).
- HCT116 p53−/− (left) cells transiently transfected with or without pcDNA4/ZBTB2‐LgBiT (ZBTB2), pcDNA4/ZBTB2 4A‐LgBiT 4A (4A), or pcDNA4/LgBiT (−) and pcDNA4/ZBTB2‐SmBiT (ZBTB2), pcDNA4/ZBTB2 4A‐SmBiT (4A), or pcDNA4/SmBiT (−), as indicated, were cultured under < 0.1% oxygen and subjected to the split luciferase complementation assay.
- Root mean square deviation (RMSD) of the backbone Cα atoms with respect to the MD‐initial structure for the native (blue) and 4A mutant (red) ZBTB2 monomer proteins.
- RMSF of the backbone Cα atoms for the native (blue) and 4A mutant (red) ZBTB2 monomer proteins.
- The same kind of luciferase assay to quantify the transactivation activity as (B) was carried out using pEF6/ZBTB2 (ZBTB2), pEF6/ZBTB2 4A (4A), or pEF6/myc‐His B (EV).
- Growth of the indicated tumor xenografts was analyzed.
Data information: Mean ± s.d. The number of technical replicates was 3 (B, D, G) and 7 (H), and reproducibility of the results was confirmed at least three times by biologically independent experiments (B–D, G, H). N.S., not significant, *P < 0.05, **P < 0.01, ****P < 0.0001, Student's t‐test (B, D, G, H).
Source data are available online for this figure.
