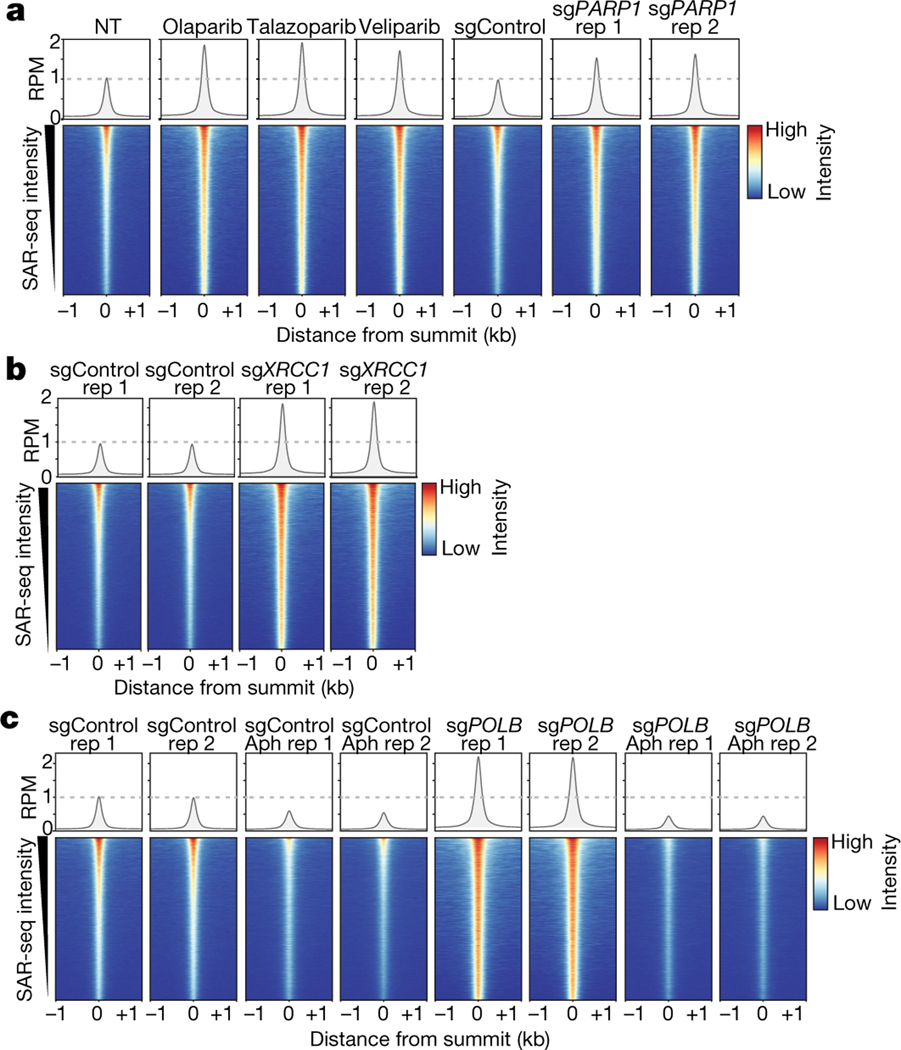
Fig. 3 |. Localized SSB repair in neurons comprises short-patch and long-patch sub-pathways.
a, Bottom, heat maps of SAR-seq intensities for 1 kb on either side of SAR-seq peak summits for i3Neurons treated with the indicated PARP inhibitors (n = 3) or expressing non-targeted control (sgControl, n = 1) or PARP1-targeted (sgPARP1, n = 2) CRISPRi plasmids. Top, aggregate plots of SAR-seq intensity. Rep, replicate. b, Heat maps of SAR-seq intensities for 1 kb on either side of SAR-seq peak summits for i3Neurons expressing non-targeted control (sgControl, n = 2) or XRCC1-targeted (sgXRCC1, n = 2) CRISPRi plasmids. c, Heat maps of SAR-seq intensities for 1 kb on either side of SAR-seq peak summits for i3Neurons expressing non-targeted (sgControl, n = 2) or POLB-targeted (sgPOLB, n = 2) CRISPRi plasmids, either untreated or treated with 50 μM aphidicolin (Aph) for 24 h before and during EdU incorporation.