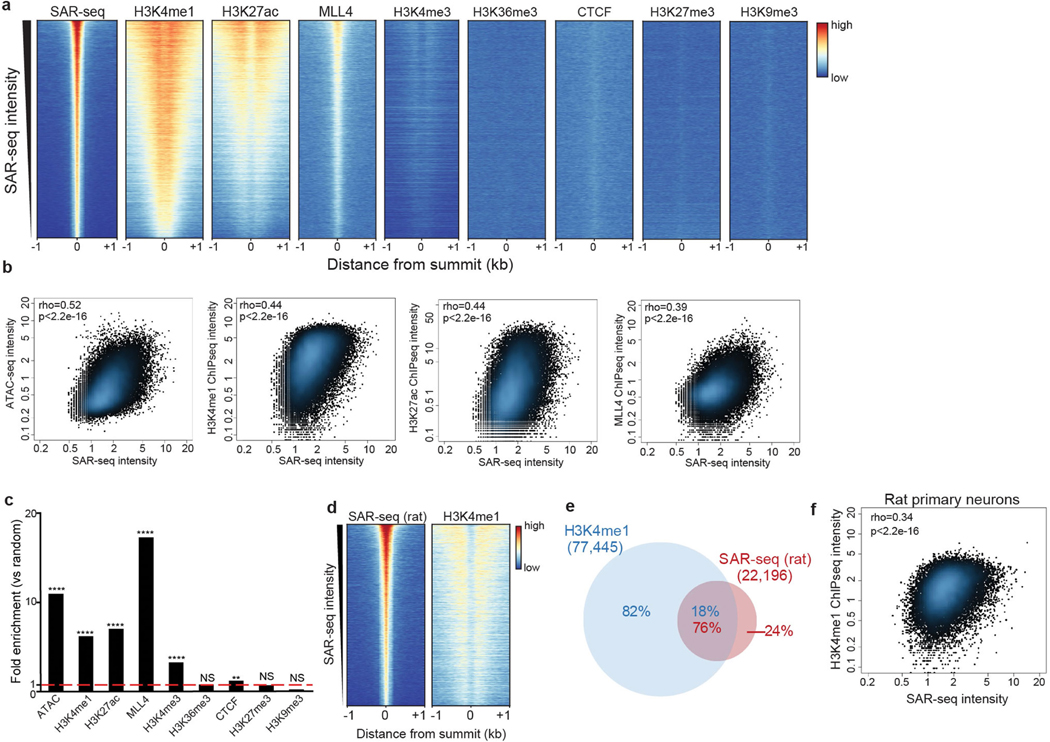
Extended Data Fig. 5 |. The correlation between SAR-seq and chromatin features.
a, Heat maps of SAR-seq and ChIP–seq data for enhancer markers (H3K4me1, H3K27ac and MLL4), other chromatin markers at accessible regions (H3K4me3, H3K36me3 and CTCF) and chromatin silencing markers (H3K27me3 and H3K9me3) for 1 kb on either side of the SAR-seq peak summit in i3Neurons, ordered by SAR-seq intensity. b, Scatter plots showing the correlation between SAR-seq intensity and ATAC–seq, H3K4me1, H3K27ac and MLL4 ChIP–seq intensities (RPKM) for 1 kb on either side of the SAR-seq peak summits in i3Neurons. Spearman correlation coefficients and P values are indicated. c, Fold enrichment of SAR-seq peaks at ATAC–seq peaks, ChIP–seq peaks of enhancer-related marks (H3K4me1, H3K27ac and MLL4), additional chromatin marks at accessible regions (H3K4me3, H3K36me3 and CTCF) (all n = 1) and chromatin silencing marks (H3K27me3 and H3K9me3) (both n = 1) in i3Neurons. n = 1,000 randomly shuffled data sets were generated to test the significance using one-sided Fisher’s exact test (P < 2.2 × 10−16 for H3K4me1, H3K27ac, ATAC–seq and MLL4; P = 1.85 × 10−316 for H3K4me3; P = 0.00116 for CTCF; **P < 0.001, ****P < 0.00001, NS, not significant). d, Heat maps of SAR-seq and H3K4me1 ChIP–seq signal for 1 kb on either side of the SAR-seq peak summit in primary rat cortical neurons, ordered by SAR-seq intensity. e, Venn diagram showing the overlap between H3K4me1 and SAR-seq peaks in rat primary neurons. n = 1,000 randomly shuffled data sets were generated to test the significance using one-sided Fisher’s exact test: P < 2.2 × 10−16. f, Scatter plot showing the correlation between SAR-seq and H3K4me1 ChIP–seq intensities (RPKM) for 1 kb on either side of the SAR-seq peak summits in rat primary neurons. Spearman correlation coefficient and P values are indicated.