Figure 1 .
Genome editing tools and the arising concerns
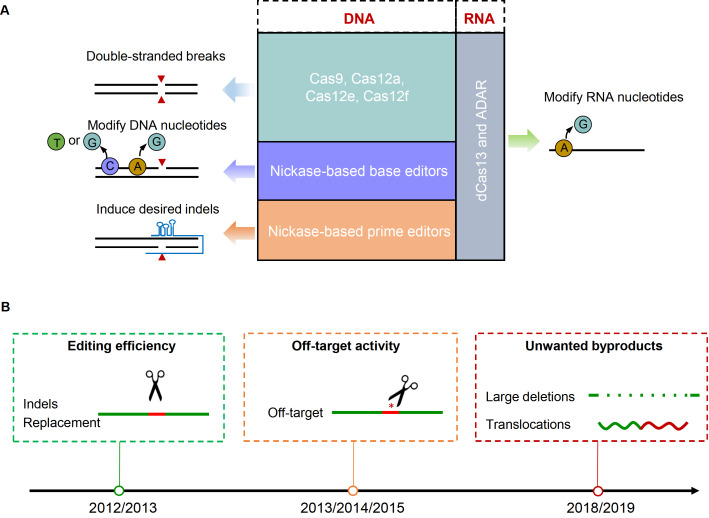
(A) Currently used CRISPR-Cas genome editing tools. CRISPR-Cas editing tools can be subtyped into DSB-dependent nucleases, nickase-based base editors, nickase-based prime editors, and dCas13-based RNA editors. Cas9, Cas12a, Cas12e, and Cas12f are widely-used nucleases for genome editing. Base editors can be classified into CBE (C to T), GBE (C to G), and ABE (A to G) based on the conversion or transversion of the nucleotides. Prime editors induce specific insertions and deletions by using an RNA template in the sgRNA scaffold. RNA base editors are designed by fusing dCas13 and ADAR to convert A to G. (B) Concerns in the genome editing field. Editing efficiencies and off-target activities are early concerns in the field. Until recently, unwanted byproducts like large deletions and translocations are appealed by NIH.