Figure1 .
Phylogenetic analysis and multiple sequence alignment of TpsAgo
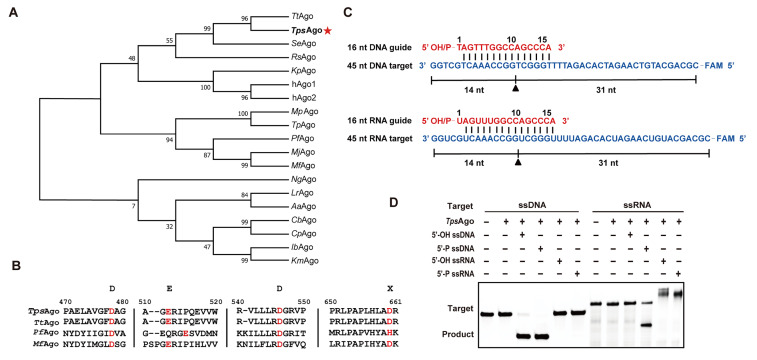
(A) Phylogenetic analysis of TpsAgo based on amino acid sequence. TtAgo: Thermus thermophilus Ago. TpsAgo: T. parvatiensis Ago. SeAgo: Synechococcus elongatus Ago. RsAgo: Rhodobacter sphaeroides Ago. KpAgo: Kluyveromyces polysporus Ago. hAgo1 and hAgo2: Homo sapiens Ago. MpAgo: Marinitoga piezophila Ago. TpAgo: Thermotoga profunda Ago. PfAgo: Pyrococcus furiosus Ago. MjAgo: Methanocaldococcus jannaschii Ago. MfAgo: M. fervens Ago. NgAgo: Natronobacterium gregory Ago. LrAgo: Limnothrix rosea Ago. AaAgo: Aquifex aeolicus Ago. CbAgo: Clostridium butyricum Ago. CpAgo: C. perfringens Ago. IbAgo: Intestinibacter bartlettii Ago. KmAgo: Kurthia massiliensis Ago. Numbers at the nodes indicate the bootstrap values for maximum likelihood analysis of 1000 resampled data sets. (B) Multiple sequence alignment of TpsAgo with several other characterized prokaryotic Agos. Red font denotes the key catalytic residues. (C) Schematic diagram of synthesized 45 nt FAM-labeled ssDNA or RNA (blue) as targets and 16 nt DNA or RNA with a 5′-OH or 5′-P group as guides (red). (D) Cleavage activity assay of TpsAgo with FAM-labeled targets.