Abstract
Bacterial protein glycosylation is commonly mediated by oligosaccharyltransferases (OTases) that transfer oligosaccharides en bloc from preassembled lipid-linked precursors to acceptor proteins. Natively, O-linking OTases usually transfer a single repeat unit of the O-antigen or capsular polysaccharide to the side chains of serine or threonine on acceptor proteins. Three major families of bacterial O-linking OTases have been described: PglL, PglS, and TfpO. TfpO is limited to transferring short oligosaccharides both in its native context and when heterologously expressed in glycoengineered Escherichia coli. On the other hand, PglL and PglS can transfer long-chain polysaccharides when expressed in glycoengineered E. coli. Herein, we describe the discovery and functional characterization of a novel family of bacterial O-linking OTases termed TfpM from Moraxellaceae bacteria. TfpM proteins are similar in size and sequence to TfpO enzymes but can transfer long-chain polysaccharides to acceptor proteins. Phylogenetic analyses demonstrate that TfpM proteins cluster in distinct clades from known bacterial OTases. Using a representative TfpM enzyme from Moraxella osloensis, we determined that TfpM glycosylates a C-terminal threonine of its cognate pilin-like protein and identified the minimal sequon required for glycosylation. We further demonstrated that TfpM has broad substrate tolerance and can transfer diverse glycans including those with glucose, galactose, or 2-N-acetyl sugars at the reducing end. Last, we find that a TfpM-derived bioconjugate is immunogenic and elicits serotype-specific polysaccharide IgG responses in mice. The glycan substrate promiscuity of TfpM and identification of the minimal TfpM sequon renders this enzyme a valuable additional tool for expanding the glycoengineering toolbox.
Keywords: O-glycosylation, oligosaccharyltransferase, pilin, bioconjugate, Moraxella osloensis
Introduction
Protein glycosylation is the most common post-translational modification and is prevalent in all 3 domains of life. In bacteria, protein glycosylation can be mediated directly by glycosyltransferases that sequentially glycosylate acceptor proteins with individual monosaccharides or by oligosaccharyltransferases (OTases) that transfer a preassembled oligosaccharide en bloc to acceptor proteins. Bacterial OTases comprise a family of glycosyltransferases that fall into 2 major categories: N-linking and O-linking (Nothaft and Szymanski 2010; Harding and Feldman 2019). N-linking OTases catalyze the covalent linkage of pre-assembled oligosaccharides to the side chains of asparagine residues in proteins, whereas O-linking OTases typically transfer glycans to the side chains of serine or threonine residues (Nothaft and Szymanski 2010). The amino acid motifs on proteins recognized by OTases are commonly referred to as sequons. Both N- and O-linking OTases are membrane-bound enzymes residing in the inner membrane and glycosylate proteins in the periplasmic space with glycans derived from lipid-linked precursors. The sugar substrate for these glycosylation reactions usually derives from a highly conserved pathway that synthesizes lipid-linked oligosaccharides on the inner leaflet of the cytoplasmic membrane that are “flipped” to the outer leaflet in a manner analogous to the Wzy-dependent pathway for O-antigen biosynthesis (Raetz and Whitfield 2002). The lipid carrier molecule for these reactions is typically undecaprenol pyrophosphate (Und-PP) and OTases catalyze the transfer of Und-PP-linked glycans to acceptor proteins. Many O-linking OTases have been shown to be promiscuous with their polysaccharide substrate and are able to transfer a variety of different bacterial glycans to proteins (DiGiandomenico et al. 2002; Faridmoayer et al. 2008). These bacterial glycans are typically composed of repeating sets of 3–6 monosaccharides known as repeat units that are polymerized in the periplasm prior to transfer (Harding and Feldman 2019). Aside from providing basic insights into bacterial glycosylation pathways, OTases have garnered significant interest due to their ability to transfer diverse bacterial O-antigen and capsular polysaccharides to specific residues on engineered carrier proteins in a process termed bioconjugation or protein–glycan coupling technology (Feldman et al. 2005; Kay et al. 2016; Harding and Feldman 2019; Kay et al. 2019). The resulting bioconjugate glycoproteins have been leveraged as glycoconjugate vaccines for a variety of bacterial pathogens with multiple candidates currently in clinical trials (GSK 2022; Johnson and Johnson 2022).
Among other substrates, O-linking OTases most notably catalyze the glycosylation of type IV pilin-like proteins (Schäffer and Messner 2017). Type IV pili are composed primarily of protein subunits called major pilins that non-covalently interact to form the fibrous structure of the pilus shaft (Giltner Carmen et al. 2012). The roles of pilin glycosylation in the bacterial lifestyle are numerous. Pilus glycosylation has been shown to aid in bacterial phage defense by blocking the binding of phage particles to surface proteins (Harvey et al. 2018). In pathogenic bacteria such as Neisseria, Pseudomonas, Burkholderia, and others, pilin glycosylation has a wide variety of effects leading to increased virulence (Yakovlieva et al. 2021) by modulating tissue adherence and invasion (Marceau et al. 1998; Nguyen et al. 2012; Willcocks et al. 2020) and by blocking the binding of complement proteins leading to immune system evasion (Tan Rommel et al. 2015). This lends importance to studies exploring the potential of bacterial glycosylation systems as novel antimicrobial and anti-virulence targets (Yakovlieva et al. 2021).
Historically, O-linking OTases have been functionally classified based on several characteristics: (i) the type(s) of substrate proteins that the OTase glycosylates, (ii) the types of glycans that the enzymes are capable of transferring, often defined by the monosaccharide or disaccharide at the saccharide reducing end (the sugar covalently linked to Und-PP), (iii) the location of the sequon in acceptor proteins, and (iv) the number of sugar monomers or oligosaccharide repeat units in the glycan that is transferred to the acceptor protein. Three major types of bacterial O-linking OTases have been previously identified and archetypal representatives from Neisseria (Faridmoayer et al. 2007), Pseudomonas (Castric 1995; Horzempa et al. 2006; Harvey et al. 2009; Qutyan et al. 2010), and Acinetobacter (Harding et al. 2015; Harding et al. 2019; Knoot et al. 2021) have been characterized. TfpO (formerly known as PilO) was the first bacterial OTase discovered and exclusively catalyzes the glycosylation of a single protein, PilA, which is the major pilin subunit of type IV pili (Giltner Carmen et al. 2012). TfpO was first discovered in Pseudomonas aeruginosa strain 1244 (P. aeruginosa 1244) (Castric 1995; Comer Jason et al. 2002) but, more recently, TfpO orthologs from other organisms have been identified in medically relevant Acinetobacter species (Harding et al. 2015). TfpO proteins catalyze the attachment of a single O-antigen repeat unit to the C-terminal serine residue on PilA both in the native organism (Castric et al. 2001) and when heterologously expressed in E. coli (Faridmoayer et al. 2007). PglL (also referred to as PglO) proteins are another class of bacterial O-linking OTases and are considered “general” OTases, catalyzing the glycosylation of multiple periplasmic and membrane-associated proteins including the major pilin subunit PilE of Neisseria type IV pili (Faridmoayer et al. 2007; Vik et al. 2009; Hayes et al. 2021). PglL orthologs naturally transfer glycans with either 2-N-acetyl sugars or galactose at the reducing end of the glycan (Faridmoayer et al. 2008), but do not transfer glycans with glucose as a reducing end sugar (Harding et al. 2019). PglS orthologs are the third type of O-linking bacterial OTase and are the most recently discovered of the 3 types (Harding et al. 2015). PglS from Acinetobacter baylyi ADP1 (PglSADP1) is the most well-characterized and natively catalyzes the glycosylation of a single pilin protein called ComP that forms type IV-like pili involved in natural competence (Porstendörfer et al. 2000; Harding et al. 2015). Notably, PglSADP1 has the broadest substrate scope of all OTases studied so far, transferring virtually any glycan to ComP including those having 2-N-acetyl sugars, galactose, or glucose at the reducing end (Harding et al. 2019).
Notwithstanding differences in sequence and ostensibly protein structure, there are several notable distinctions between TfpO, PglL, and PglS proteins. First, TfpO proteins glycosylate the C-terminal serine or threonine residue of substrate proteins (Comer Jason et al. 2002), whereas PglL and PglS proteins glycosylate sequons that are in the interior of the acceptor protein coding sequence (Vik et al. 2009; Harding et al. 2019). Second, TfpO proteins are limited to transferring only short oligosaccharides to acceptor proteins, while PglL and PglS proteins, when heterologously expressed in glycoengineered E. coli systems, can transfer long-chain polysaccharides with multiple repeat units composed of possibly hundreds of monosaccharides (Comer Jason et al. 2002; Horzempa et al. 2006; Faridmoayer et al. 2007; Harding et al. 2015, 2019). However, the reason for this difference in glycan substrate size and reducing-end sugar preference are not known, in part due to the unavailability of protein structures of any bacterial O-linking OTases.
In this work we describe a new, 4th class of O-linking OTase carried by bacteria from the Moraxellaceae family that shares properties with both TfpO and PglS proteins. These new OTases, herein referred to as TfpM, are closer in length and amino acid sequence similarity to TfpO proteins yet can transfer long-chain polysaccharides like PglS and PglL proteins when expressed in E. coli. After initial screening of several representatives from this new class of O-linking OTase, we focused our detailed characterization on a TfpM ortholog from M. osloensis termed TfpMMo. First, we identified the site of glycosylation on an engineered pilin-fusion protein using site-directed mutagenesis and mass spectrometry. TfpM-mediated pilin glycosylation was also confirmed in the native M. osloensis bacteria using glycoproteomics. Using a reductive cloning strategy, we were able to determine the minimum number of amino acid residues from the native pilin substrate required for glycosylation by TfpMMo and investigated the different types of polysaccharide substrates that TfpMMo can transfer. Interestingly, TfpMMo was found to have a broad glycan substrate range like PglS and can transfer diverse capsular and O-antigen polysaccharides from a variety of bacteria including Salmonella, Streptococcus pneumoniae, Klebsiella pneumoniae, and Group B Streptococcus (GBS). Additionally, we demonstrate that a bioconjugate generated by TfpM containing the type III capsular polysaccharide from GBS is immunogenic, eliciting serotype III-specific IgG responses. The discovery of a new class of O-linking OTase furthers our understanding of the mechanisms that underly glycan specificity by these and related OTases and enables more comparative studies of this important enzyme family. Furthermore, the wide range of polysaccharide substrates transferred by TfpMMo, including those that have glucose as the reducing end sugar, enable this enzyme to be used to a new tool for protein glycoengineering.
Results
Bioinformatic identification of a new class of OTase carried by bacteria from the Moraxellaceae family
Given that bacteria from the genus Acinetobacter have been shown to carry genes encoding for TfpO, PglL, and PglS orthologs, we were interested in searching for new O-linking OTases within the Moraxellaceae family to which Acinetobacter belongs. As the different types of O-linking OTases cannot be clearly differentiated based on primary amino acid sequence alone, definitive family assignment typically relies on functional assays. Nevertheless, some heuristics can aid in the initial classification of OTases. For instance, TfpO proteins are typically smaller in size, around 420–460 amino acids, than PglS and PglL proteins that are approximately 525–600 amino acids. Moreover, the amino acid sequences of the pilins that O-linking OTases glycosylate are also sufficiently different between OTase families and can be used to help identify to which family an OTase belongs. In the case of PglS only a single protein, the type IV pilin-like protein ComP, serves as a cognate acceptor in its natural context (Harding et al. 2015). The PglS sequon is flanked by 2 cysteine residues and contains a conserved serine residue at position 84 that is the site of glycosylation (Harding et al. 2019; Knoot et al. 2021). In contrast, the minimum TfpO sequon is located at the C-terminus of the type IV pilin protein PilA (Comer Jason et al. 2002; Horzempa et al. 2006; Harvey et al. 2009).
To identify genes encoding O-linking OTases, we first searched the NCBI genome and whole-genome shotgun contig sequence databases using the Basic Local Alignment Search Tool (BLAST) and the PglSADP1 amino acid sequence as the query. To shorten the list of hits and reduce the likelihood of identifying very similar orthologs of PglSADP1, we further refined our search to candidates that had less than 50% amino acid sequence identity to PglSADP1. Top hits from this refined list were proteins that were much more similar in size to TfpO proteins, but the upstream cognate pilin protein contained both a ComP disulfide-flanked sequon as well as a PilA-like sequon at the C-terminus. The first identified pilin-OTase pairs were encoded in 2 Acinetobacter species: Acinetobacter parvus DSM 16617 and Acinetobacter townerii ZZC-3 (Table 1). Intrigued by these findings, we looked further and identified other species in the Moraxellaceae family encoding similar putative OTase/pilin pairs. While we found many genes like those in A. parvus DSM 16617 and A. townerii ZZC-3, most of the associated pilin proteins encoded upstream of the OTase lacked a ComP-like sequon. The accession numbers and protein sizes for 20 of these putative pilin-OTase pairs are listed in Table 1. We did not observe any homologues in species outside of the Moraxellaceae family and, to distinguish them from other known OTases, we have termed them TfpM proteins. Given the similar size of TfpM proteins to known TfpO proteins, we initially hypothesized that these genes encoded variants of TfpO-PilA pairs like those found in Acinetobacter and Pseudomonas (Harding et al. 2015). However, a multiple sequence alignment of the 20 TfpM proteins with known PglS, PglL, and TfpO proteins demonstrated that the former had less than 26% sequence identity to archetypal OTases. Analysis of the phylogenetic tree produced by the multiple alignment showed that TfpM proteins cluster in distinct clades from TfpO, PglS, and PglL proteins (Fig. 1A and Supplementary Fig. 1). In contrast, the pilin genes located immediately upstream of the tfpM genes did not cluster in discrete clades (Supplementary Fig. 2) and displayed overall higher identity to PilA proteins, namely between 37% and 60%. Based on the coding sequence, most of the associated pilin proteins belong to the type IV major pilin family with the exception of those from Acinetobacter sp. CIP102143 and Acinetobacter sp. CIP102637, which lack the characteristic type III signal sequence at the N-terminus (Giltner Carmen et al. 2012).
Table 1.
Organisms and NCBI accession numbers for the TfpM enzymes and their associated pilins.
| Organism | OTase accession number | OTase length | Pilin accession number | Pilin length |
|---|---|---|---|---|
| Acinetobacter terrae strain ANC 4282 | WP_171532559 | 422 | WP_171532558 | 150 |
| Acinetobacter johnsonii strain S00071 | WP_184117984 | 424 | WP_184117983 | 145 |
| Psychrobacter piscatorii T-3-2 | WP_201599090 | 427 | WP_201599046 | 146 |
| Psychrobacter alimentarius strain A3K91 | AMT95940 | 439 | AMT95939 | 146 |
| Psychrobacter sp. 72-O-c | WP_201554594 | 440 | WP_201554596 | 148 |
| Psychrobacter sp. BI730 | WP_179798140 | 431 | WP_179798138 | 150 |
| Acinetobacter pseudolwoffii Acilw_F78 | WP_074383624 | 424 | WP_074383625 | 142 |
| Acinetobacter parvus DSM 16617 | WP_004683394 | 422 | WP_004683396 | 150 |
| Acinetobacter towneri strain ZZC3–9 | WP_194559050 | 422 | WP_194559049 | 149 |
| Acinetobacter indicus strain AI40 | WP_168379910 | 425 | WP_168379911 | 147 |
| Acinetobacter indicus strain AI7 | WP_151711300 | 424 | WP_168416595 | 144 |
| Acinetobacter sp. YZS-X1–1 | WP_034169718 | 425 | WP_034169719 | 146 |
| Acinetobacter sp. CIP 102637 | WP_004769912 | 425 | WP_004797623 | 142 |
| Acinetobacter sp. CIP 102159 | WP_004757006 | 425 | WP_004757004 | 141 |
| Acinetobacter sp. YH01026 | WP_180158749 | 425 | WP_180158748 | 168 |
| Acinetobacter junii strain 65 | WP_075696034 | 424 | WP_075696033 | 148 |
| Acinetobacter venetianus VE-C3 | WP_019385507 | 424 | WP_019385508 | 145 |
| Acinetobacter sp. CIP 102143 | WP_005328296 | 425 | WP_005328298 | 132 |
| A. junii strain TUM15069 | WP_151711300 | 425 | WP_151711301 | 168 |
| Moraxella osloensis strain FDAARGOS_1202 | WP_062331497 | 425 | WP_156627541 | 167 |
Fig. 1.
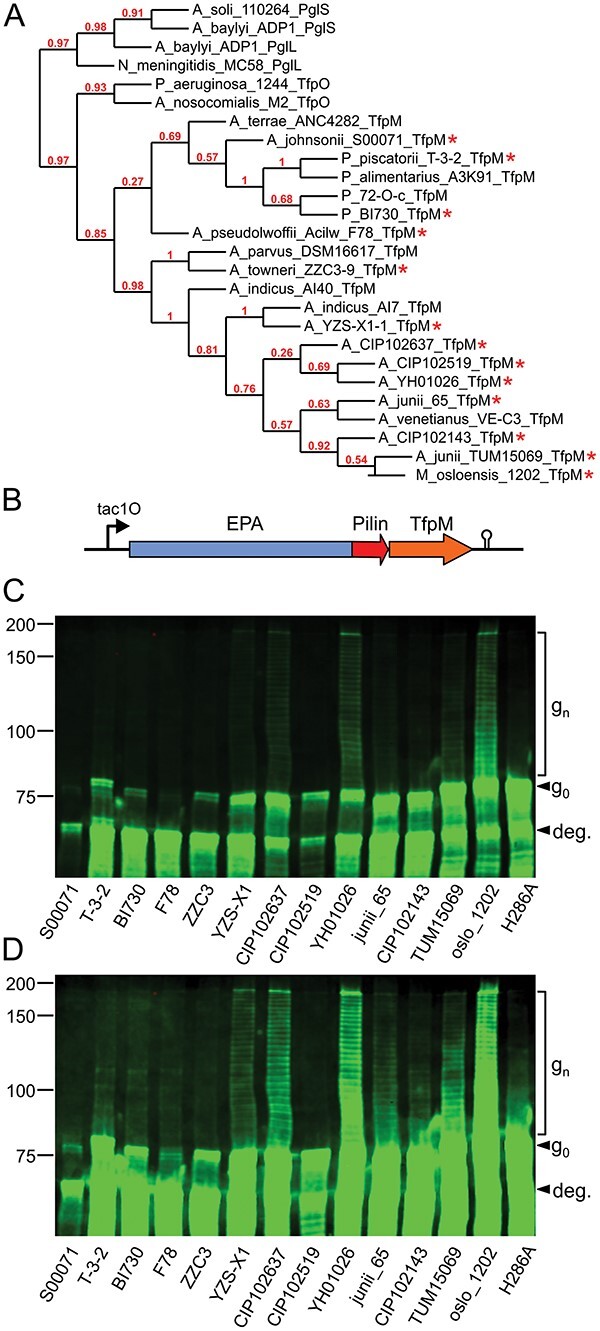
Characterization of 13 OTases (TfpM orthologs) from species in the Moraxellaceae family. A) Cladogram of the 20 TfpM orthologs with archetypal TfpO, PglL, and PglS from Acinetobacter, Neisseria, and Pseudomonas, respectively. Branch confidence is indicated in red. Starred OTase/pilin pairs were cloned and tested in bioconjugation experiments. B) Diagram showing the EPA-pilin fusion protein and TfpM construct design. Colored arrows indicate genes. Gene expression was driven from an IPTG-inducible tac promoter with a lacO operator (tac1O). The rrnB T2 terminator is marked by a black hairpin structure. C and D) Anti-EPA western blots of whole-cell E. coli extracts expressing different EPA-pilin carrier open reading frames and tfpM OTase genes. Panel D shows the same image as panel C but at higher exposure. Whole cell extracts loaded in each lane were normalized by OD600. H286A indicates the Moraxella osloensis TfpM site directed OTase active site mutant. “g0” indicates unglycosylated EPA-pilin and “gn” indicates CPS8-glycosylated EPA-pilin protein. “deg.” indicates partially degraded carrier protein. Reference protein masses are marked left of the western blot in kDa.
TfpM orthologs glycosylate engineered pilin-fusion proteins with the pneumococcal serotype 8 capsular polysaccharide
Although similar in size to TfpO proteins, TfpM proteins are sufficiently different in terms of their amino acid sequence to warrant further investigation. Of the 20 TfpM OTases listed in Table 1, we selected 13 representatives from different clades to test for glycosylation activity in a glycoengineered E. coli strain (Feldman et al. 2005). Previously, we developed a chimeric acceptor protein strategy consisting of the exotoxin A protein from P. aeruginosa (EPA) fused to different-sized, soluble fragments of ComP (the natural substrate of PglS) (Harding et al. 2019; Knoot et al. 2021). All type IV pilin-like proteins contain a conserved, N-terminal pilin signal sequence, and membrane-anchoring domain that is not required for glycosylation and is thus deleted in EPA-pilin fusion proteins. We used this approach to screen the TfpM-pilin pairs and designed 13 synthetic double-stranded DNA blocks encoding an N-terminally truncated fragment of the upstream pilin gene and the downstream tfpM gene. The truncated pilin fragments ranged from 113 to 140 amino acids and were cloned in-frame with the upstream EPA coding sequence (Fig. 1B). No purification tag was added to the C-terminus of the EPA-pilin fusion as previous studies with TfpO from P. aeruginosa 1244 reported that adding additional C-terminal residues after the serine residue prevented glycosylation (Horzempa et al. 2006). Expression of the EPA-pilin and TfpM proteins was driven from an IPTG-inducible tac promoter on a pEXT20 plasmid (Dykxhoorn et al. 1996). The fusion protein was secreted into the periplasm using a DsbA signal sequence at the N-terminus of EPA (Fig. 1B). Oligos and primers used for the assemblies are listed in Supplementary Table 1.
We assessed the ability of the 13 TfpM proteins to transfer the S. pneumoniae capsular polysaccharide 8 (CPS8) glycan to their cognate pilin domain on the EPA-pilin fusions. The CPS8 repeat unit is a tetrasaccharide with a glucose at the reducing end. Notably, PglS is thus far the only known OTase able to naturally transfer this glycan to acceptor proteins (Harding et al. 2019). The 13 EPA-pilin fusions/TfpM expression vectors were transformed individually into E. coli SDB1 strains (Feldman et al. 2005) expressing the CPS8 glycan and assessed for protein glycosylation. The unglycosylated fusion proteins had expected masses ranging from 78.3 to 80.5 kDa. Several TfpM proteins were found to glycosylate their cognate EPA-pilin fusion with glycosylation appearing as higher molecular-weight laddering (gn) above the unglycosylated band (g0) (Fig. 1C and D). Each higher-weight band represents the attachment of a glycan with 1 additional CPS8 repeat unit to the EPA-pilin protein. Glycosylation was observed in 7 TfpM orthologs tested: Acinetobacter sp. YZSX-1-1, Acinetobacter sp. CIP102637, Acinetobacter sp. YH01026, Acinetobacter junii 65, Acinetobacter sp. CIP102143, Acinetobacter sp. TUM15069, and M. osloensis FDAARGOS_1202 (Fig. 1C). A Wzy_C protein domain is present in all known PglS, PglL, TfpO, and TfpM orthologues and, as a control, we generated a mutant of the M. osloensis TfpM protein gene with a single-residue change in a conserved histidine (His286) previously shown to be required for activity of wzy_C “O-antigen ligase” domain-containing enzymes (Supplementary Fig. 3) (Ruan et al. 2012; Musumeci et al. 2014). We did not observe any glycosylation in SDB1 cell extracts expressing the H286A mutant alongside the EPA-pilin fusion protein and CPS8 glycan (Fig. 1C and D).
We observed that most of the EPA-pilin constructs were poorly expressed and/or prone to degradation. Degradation is apparent in every lane from an intense protein band running at lower weight near 70 kDa as well as multiple less-intense bands below the expected intact mass (“deg.” in Fig. 1C and D). The expected mass of EPA alone is 66.6 kDa. We have previously observed similar instability of EPA fusion constructs with ComP and hypothesized that this was due to proteolytic degradation of the truncated C-terminal pilin domain (Knoot et al. 2021). Since some of the pilins were from potentially psychrophilic Psychrobacter species, we hypothesized that the instability of the EPA-pilins could be mitigated by lowering the temperature. However, glycosylation experiments performed with these strains at 20 °C rather than 30 °C still resulted in similar levels of protein degradation (data not shown).
TfpMMo is an O-linking OTase that glycosylates the C-terminal threonine of its pilin substrate
Of the 13 TfpM-pilin pairs tested in the preceding experiment, those from M. osloensis 1202 and Acinetobacter sp. YH01026 resulted in the most effective transfer of glycans of diverse sizes. Due to the slightly higher apparent stability of the pilin from M. osloensis FDAARGOS_1202 (1202 hereafter), we chose the OTase from this organism as a representative for further characterization and have termed the enzyme TfpMMo. For clarity, we refer to the intact, native M. osloensis 1202 pilin protein as PilMo and the N-terminally truncated fusion domain as PilMoΔ28 throughout the text as it is missing its first 28 amino acids. Using TfpMMo, we next aimed to identify the glycosylation site of PilΔ28 and thereby determine whether the enzyme acted like a TfpO protein, glycosylating the C-terminal amino acid of its cognate pilin acceptor, or if it was more like a PglL or PglS protein, glycosylating an internal residue. All except one of the cognate pilin proteins located immediately upstream a tfpM open reading frame ended in a C-terminal threonine residue, namely that from Psychrobacter sp. 72-O-c that ends in a serine. Based on this observation, we hypothesized that TfpM enzymes transfer glycans to a C-terminal residue and thus generated 2 mutants of the C-terminal pilin threonine (Thr167), converting this residue to either serine or alanine and tested for glycosylation with CPS8. Whole cell E. coli extracts of the strains expressing the PilMoΔ28 mutants were probed using anti-EPA antibody in western blots. As seen in Supplementary Fig. 4, TfpMMo was also able to glycosylate EPA-PilMoΔ28 T167S but not the alanine mutant. These results indicate that the hydroxyl group on the side chain of the Thr167 or Ser167 residue is likely the site of glycan attachment by TfpMMo.
To confirm the site of pilin glycosylation by TfpMMo, we partially purified CPS8-glycosylated EPA-PilMoΔ28, separated it via SDS-PAGE analysis and Coomassie-stained the resolved glycoproteins. Gel slices corresponding to EPA- PilMoΔ28 glycosylated with 1–3 CPS8 repeat units were excised (Supplementary Fig. 5), digested with LysC, and analyzed for glycopeptides. Open searching-based analysis (Chick et al. 2015; Polasky et al. 2020) of LysC-derived EPA-PilMoΔ28 peptides resulted in the identification of Hexose (Hex)-Hexuronic Acid (HexA)-modified 762FLPANCRGT770 peptide consistent with an incomplete monomer of the CPS8 glycan (HexHexAHex2, Supplementary Table 2). Higher-energy C-trap dissociation (HCD) analysis supports linkage of this disaccharide through the hexose residue and targeted Electron-Transfer/Higher-Energy Collision Dissociation (EThcD) analysis confirmed the attachment of HexHexA to the C-terminal threonine residue (Fig. 2A and B). While this MS analysis did not identify multimers of the CPS8 tetrasaccharide this is not surprising due to elongated glycoconjugates being extremely difficult to detect using peptide-centric liquid chromatography–mass spectrometry (LC–MS) methods without the use of specialized chemical additives such as supercharging agents (Lin et al. 2016). Nevertheless, the identification of a disaccharide consistent with the partially completed CPS8 tetrasaccharide still supports the identity of the high molecular weight laddering as polymerized CPS8 tetrasaccharides.
Fig. 2.

Targeted MS/MS analysis of the HexHexA-modified C-terminal EPA-Pil∆28 peptide 762FLPANCRGT770. A) EThcD fragmentation enabled the localization of the HexHexA glycosylation event to the terminal residue Thr770 of EPA-Pil∆28. B) HCD fragmentation enables the confirmation of the peptide sequence as well as the linkage of the disaccharide HexHexA through the Hex monosaccharide by the observation of multiple y-ions linked solely to a Hex residue.
M. osloensis CCUG 350 glycosylates PilMo at the C-terminal threonine with multiple glycan compositions
The observation that TfpM glycosylates the C-terminal threonine of PilMoΔ28 in the context of a glycoengineered E. coli system supports the notion that TfpM possesses OTase activity unique to those previously characterized. To further understand this activity, we examined PilMo glycosylation within its native context using high-field asymmetric waveform ion mobility spectrometry (FAIMS)-based glycoproteomic analysis of M. osloensis. Sequence comparisons of the M. osloensis 1202 draft genome with other publicly accessible strains identified M. osloensis CCUG 350 as having an identical tfpM sequence and associated pilin gene sequence to that in M. osloensis 1202; as such, we used the M. osloensis CCUG 350 strain for FAIMS-based glycoproteomic analysis. Analysis of MS/MS events in M. osloensis CCUG 350 corresponding to the C-terminal PilMo peptide 159FLPANCRGT167 revealed 3 different glycans decorating PilMo (Fig. 3 and Supplementary Fig. 6): dhexNAc-dhex-hex-hex-dhex (803.321 Da, Fig. 3A and B), dhexNAc-dhex-hex-hex-dhex-Hex (965.375 Da, Fig. 3C and D), and hexNAc-hex-hex-317 (843.3128 Da, Fig. 3E and F). HCD and EThcD confirmed the identity of these glycopeptides and their attachment to the terminal threonine residue (Fig. 3). The 317 Da residue in hexNAc-hex-hex-317 is consistent with legionaminic/pseudaminic acid commonly observed within Acinetobacter baumannii glycoproteomes (Scott et al. 2014). Interestingly, open database searching of M. osloensis CCUG 350 also revealed non-PilMo-associated peptides decorated with a mass of 843.31 Da (Supplementary Table 3 and Supplementary Fig. 7) supporting the presence of a second, general O-linking glycosylation system in addition to the TfpM system characterized here. This non-PilMo glycosylation event may be mediated by a putative PglL-like OTase encoded in gene locus AXE82_RS03530 that is located near the tfpM and pilin-associated genes in M. osloensis CCUG 350.
Fig. 3.
Multiple glycans are observed decorating the PilMo from Moraxella osloensis CCUG 350. EThcD and HCD MS/MS analysis demonstrate the C-terminal peptide 159FLPANCRGT167 can be modified on the terminal Threonine residue with the glycans dhexNAc-dhex-hex-hex-dhex (803.321 Da, A and B), dhexNAc-dhex-hex-hex-dhex-Hex (965.375 Da, C and D) and HexNAc-Hex-Hex-317 (843.3128 Da, E and F).
TfpMMo transfers polysaccharides containing glucose, galactose, or 2-N-acetyl monosaccharides at the reducing end
Next, we sought to explore the range of polysaccharide substrates that TfpMMo can transfer in glycoengineered E. coli backgrounds. We selected polysaccharides containing different reducing-end sugars, varying disaccharide sugar linkages near the reducing end, and/or were polymers composed of linear or branched repeat units. In addition to pneumococcal CPS8, 4 polysaccharides were tested as glycan substrates for TfpMMo: E. coli O16 antigen, Salmonella enterica LT2 O-antigen, K. pneumoniae O2a antigen, and the type III Group B Streptococcus capsular polysaccharide (GBSIII). The structures of all 5 repeat units tested are shown in Fig. 4A (Whitfield et al. 1992; Curd et al. 1998; Pinto and Berti 2014; Geno et al. 2015; Liu et al. 2020). Each of the 5 polysaccharides were individually co-expressed with EPA-PilMoΔ28 and TfpMMo in E. coli SDB1 cells, induced, and grown for subsequent glycoprotein purification. Periplasmic extractions of SDB1 cells were partially purified using anion-exchange chromatography to remove any contaminating Und-PP-linked polysaccharides that would convolute interpretation of the western blots. The western blots were probed using antisera specific to each polysaccharide and, separately, with anti-EPA antibody as all antibodies used in this experiment were from rabbits. As seen in Figure 4, TfpMMo was found to transfer all 5 different polysaccharides to the EPA-PilMoΔ28 protein. Digestion of the bioconjugates with proteinase-K prior to western blotting eliminated both the anti-glycan (Fig. 4B) and anti-EPA (Fig. 4G) signals, confirming that the anti-glycan signals originated from protein-linked rather than contaminating lipid-linked polysaccharide.
Fig. 4.
TfpMMo can transfer diverse bacterial glycans to the EPA-PilMoΔ28 carrier protein. A) Structures of the repeat units of the 5 bacterial glycans tested with TfpMMo. The linkages between sugar monomers are indicated in rounded brackets. Glycan abbreviations used: CPS8, Streptococcus pneumoniae capsular polysaccharide 8; GBSIII, Group B Streptococcus capsular polysaccharide III; LT2, Salmonella enterica Group B serotype LT2 O-antigen; O16, E. coli serotype O16 O-antigen; O2a, Klebsiella pneumoniae serotype O2a O-antigen; All sugars are the pyranose form except where noted. Abbreviations used: Glc, glucose; Gal, galactose; Galf, galactofuranose; Rha, rhamnose; GlcNAc, N-acetylglucosamine; Abe, abequose; NeuNAc, N-acetylneuraminic acid, sialic acid. B–F) Anti-glycan western blots with partially purified TfpMMo-derived bioconjugates. B) Anti-CPS8. C) Anti-O16. D) Anti-LT2. E) Anti-O2a. F) Anti-GBSIII. G) Anti-EPA. In panels B–G, ± labels indicate whether the samples were incubated with (+) or without (−) proteinase K prior to SDS-PAGE separation. Reference protein masses are marked next to the western blots in kDa.
TfpMMo can glycosylate truncated PilMoΔ28 variants
The prior experiments all utilized an N-terminally truncated variant of PilMo that was 139 amino acids in length. To gain insights into the minimal features required for C-terminal pilin glycosylation by TfpMMo, we generated a series of further-truncated variants of PilMoΔ28 and tested whether they could be glycosylated. We first generated a 20 amino acid fragment of PilMo fused C-terminally to EPA via a flexible 4-residue glycine linker, termed Pil20 (Fig. 5A). This 20-amino acid fragment was chosen because it contains a disulfide loop (“DSL”) region that is conserved in many type IV pilins (Supplementary Fig. 8) (Horzempa et al. 2006; Harvey et al. 2009). Based on sequence alignments with P. aeruginosa 1244 PilA, the DSL in M. osloensis pilin is formed by residues Cys148 and Cys164 and we designed the sequence downstream of the glycine linker to start with Cys148. The plasmid encoding EPA-Pil20 and TfpM was termed pVNM297. Glycosylation experiments with Pil20 revealed that it was able to be glycosylated by TfpMMo with CPS8 at similar levels to PilMoΔ28 (Fig. 5B). To test whether the DSL region was required for glycosylation, we generated several shorter variants that lacked part of this feature. In these smaller constructs, we also mutated Cys164 to alanine to prevent the formation of non-natural disulfide linkages upon oxidation in the periplasm (Harvey et al. 2009). We tested 15-, 13-, and 10-amino acid PilMo variants, with one variant of the 10-amino acid version containing a GGGG linker and one without. These constructs were termed Pil15, Pil13, Pil10L, and Pil10, respectively (Fig. 5A). TfpMMo was able to glycosylate all 4 of these variants, albeit at lower levels than Pil20 and PilMoΔ28 (Fig. 5B). Both Pil10 and Pil10L were glycosylated comparably, indicating that the presence of the upstream glycine linker does not have a marked effect on TfpMMo activity. This linker was omitted from all subsequent constructs. Of the 4 proteins, the Pil13 was glycosylated considerably worse than the others.
Fig. 5.

TfpMMo glycosylates truncated EPA-fused pilin variants as small as 3 amino acids. A) Sequences of the EPA-fused PilMo fragments tested for bioconjugation with TfpMMo. Blue letters mark the C-terminal residues of EPA. Underlined residues indicate the glycine linker placed between EPA and the pilin sequence. B) Anti-EPA western blot of whole cell extracts expressing the truncated pilin variants, CPS8, and TfpMMo. The calculated EPA-PilMO∆28 mass is 80.3 kDa and that of the truncated variants ranges from 67.1 to 69.0 kDa. All lanes were normalized to the same OD600. “g0” indicates the unglycosylated truncated EPA-pilins and “gn” indicates the glycosylated EPA-pilin protein. Unglycosylated EPA-PilMo∆28 runs near 75 kDa. Reference protein masses are marked left of the western blot in kDa.
From a sequence alignment, we noted that each of the pilin proteins that were glycosylated by TfpM OTases have a conserved “P-A-N/E-C-R-G” motif found near the C-terminus, immediately upstream of the penultimate threonine residue (Supplementary Fig. 8). Given the presence of this feature in all glycosylated pilins, we asked whether this motif was required for glycosylation by TfpMMo. We fused a 7-amino acid variant, termed Pil7, of PilMo consisting of a similar motif (modified to “P-A-N-A-R-G-T,” where the cysteine is mutated to an alanine—bolded residue) to EPA and assessed glycosylation (Fig. 5A). We also built stepwise single-amino acid truncations of this “P-A-N-A-R-G-T” sequence from 7 to 2 amino acids and assessed the ability of TfpM to glycosylate these fragments with CPS8. The results showed that all variants except for Pil2 were glycosylated by TfpMMo at similar levels as Pil10 (Fig. 5B). The overall glycosylation was again lower than for the Pil20 and PilMoΔ28. Glycosylated Pil2 was barely detectable though some trace laddering was visible at higher exposure of the western blot. This indicates that TfpMMo can glycosylate this variant but at markedly reduced levels than the construct with even 1 additional amino acid.
Immunogenicity of a TfpMMo-derived GBSIII bioconjugate
Given that the EPA-Pil20 construct was the most efficiently glycosylated of the truncated PilMoΔ28 variants by TfpM, we next assessed the immunogenicity of a EPA-Pil20 protein glycosylated GBSIII in a murine vaccination model. To aid in protein purification for these experiments, we constructed a plasmid expressing an N-terminally 6x-His tagged variant of the EPA-Pil20 carrier protein (pVNM291). The His tag was added immediately downstream of the predicted N-terminal DsbA signal sequence cleavage site of EPA. pVNM291 was introduced into SDB1 cells expressing the GBSIII glycan and resulting bioconjugates purified using Nickel immobilized metal affinity chromatography (IMAC) followed by anion-exchange and size-exclusion chromatography on Fast Protein Liquid Chromatography (FPLC). Western blot and Coomassie staining of the SDS-PAGE resolved EPA-Pil20-GBSIII bioconjugate confirmed high molecular weight glycosylation of the EPA-Pil20 protein with the GBSIII glycan (Fig. 6A–D). Intact protein MS of purified EPA-Pil20-GBSIII supported a glycan:protein ratio of 20% (Fig. 6E) calculated using methods reported in Duke et al. (2021). Each dose was formulated to contain 1 μg of GBSIII polysaccharide. As a control for these experiments, we purified unglycosylated pVNM291-derived EPA-Pil20 from SDB1 cells without glycan plasmid and dosed at the same protein concentration as for the GBSIII bioconjugates.
Fig. 6.

Purified TfpMMo-derived EPA-Pil20-GBSIII bioconjugate elicits a robust IgG immune response in mice. A) Western blot of purified EPA-Pil20-GBSIII bioconjugate, anti-EPA channel. B) Anti-GBSIII. C) Merged image of A and B. D) Coomassie stain of purified EPA-Pil20-GBSIII bioconjugate. 0.75 and 5 μg protein were loaded per lane in panels A–D, respectively. Some residual unglycosylated and partially degraded EPA-Pil20 protein is observed below 75 kDa. E) MS1 spectrum of intact, purified GBSIII-291 bioconjugate. EPA-Pil20 has a theoretical mass of 69582.19 Da. EPA-Pil20 is observed in multiple states of increasing mass separated by ~980 Da, which corresponds to the calculated mass of a GBSIII glycan repeat unit. F) GBSIII-specific IgG kinetics over the course of immunization as measured by ELISA and converted to ng/mL IgG using a standard IgG curve. **P < 0.01. Reference protein masses in kDa are marked on the left in panels A–D.
To test whether the TfpMMo-generated EPA-Pil20-GBSIII bioconjugate was immunogenic, we immunized 5-week-old female CD-1 mice. Mice received either unglycosylated EPA-Pil20 carrier protein as a control or EPA-Pil20-GBSIII at 2-week intervals starting with a priming dose followed by 2 booster doses. All vaccines were formulated with a 1:9 ratio of Alhydrogel 2% as an adjuvant. Serum was collected prior to each immunization and 2 weeks after the final booster. To determine the level of GBSIII-specific antibodies elicited, we used an enzyme-linked immunosorbent assay (ELISA). All mice immunized with the bioconjugate EPA-Pil20-GBSIII were observed to express high levels of anti-GBSIII IgG antibodies, excluding a single mouse that exhibited a low anti-GBSIII IgG responses but was able to be boosted over the course of immunization (Fig. 6F). As expected, the EPA-Pil20-GBSIII-vaccinated mice had increased GBSIII-specific IgG titers compared with the control EPA-Pil20-vaccinated mice (EPA-Pil20, Fig. 6F). Collectively, these data suggest that TfpMMo can produce bioconjugates capable of eliciting polysaccharide-specific IgG responses.
TfpMMo and PglSADP1 glycosylate a single protein engineered to contain sequons specific to each OTase
Lastly, we wanted to determine if a protein engineered to contain sequons from 2 different OTase systems could be glycosylated by both OTases at each site. As such, we constructed an EPA fusion protein containing a sequon associated with TfpM as well as a sequon associated with PglS. To this end, we engineered an EPA fusion protein containing a PglS sequon (CTGVTQIASGASAATTNVASAQC) integrated between residues Ala489 and Arg490 as previously described (Knoot et al. 2021) as well as the Pil20 sequon (CGGTGTTVAAKFLPANCRGT) at the C-terminus (Fig. 7A) with tfpM immediately downstream. The pglS gene from A. baylyi ADP1 was cloned 100 bp downstream of the tfpM stop codon. We introduced this vector (pVNM337) into E. coli SDB1 expressing the E. coli O16 antigen and tested for glycosylation via western blotting. To compare against proteins containing only a single sequon, we individually introduced the following constructs into E. coli SDB1 expressing the O16 antigen: (i) EPA containing only the TfpM-associated Pil20 sequon (pVNM297) or (ii) EPA containing the PglS sequon integrated between residues Ala489 and Arg490 (pVNM167). To compare against a protein that has 2 sequons and can be di-glycosylated, we expressed an EPA construct containing the PglS sequon integrated between residues Ala489 and Arg490 as well as between residues Glu548 and Gly549 (pVNM245, Fig. 7A). As seen in Figure 7B, western blot analysis of EPA constructs containing only one sequon either from ComP or PilMo exhibited a glycoprofile around 100 kDa, indicative of a single glycosylation event. The EPA construct containing 2 PglS sequons exhibited a predominantly mono-glycosylated profile around 100 kDa but also a di-glycosylated population migrating around 150 kDa. Western blot analysis of the EPA fusion containing a sequon from TfpMMo and PglSADP1 displayed both mono- and di-glycosylated populations like that seen with construct pVNM245. We conclude from these results that a carrier protein can be simultaneously glycosylated by 2 different OTase classes in one expression system.
Fig. 7.

Glycosylation of EPA constructs containing sequons from 2 different O-linking OTase systems. A) Diagrams of the plasmid-based operons expressing EPA with PglS- or TfpM-specific sequons. Internal glycotag (“iGT”) is a 23 amino acid fragment derived from A. soli CIP 110264 ComP inserted between EPA Ala489–Arg490 and/or Glu548–Gly549. B) Anti-EPA western blot of SDB1 periplasmic extracts expressing one of the 4 constructs and the E. coli O16 O-antigen. Load amounts per lane were normalized to OD600. “g0” indicates unglycosylated EPA carrier protein and the singly or doubly glycosylated EPA proteins are noted. Reference protein masses are marked left of the western blot in kDa.
Discussion
Herein we describe the identification and characterization of a new family of O-linking OTases. TfpMMo represents, to our knowledge, the first characterized OTase from the Moraxella genus. The phylogenetic tree of the bacterial OTases indicates that the TfpM proteins cluster outside of the clades with known OTases such as TfpO, PglL, and PglS (Fig. 1A). The TfpM proteins described here share at least 50% sequence identity with one another but are only 15–26% identical at the amino acid level to the enzymes from Neisseria, Pseudomonas, and Acinetobacter. Due to the instability of some of the pilin fusion proteins tested, we were unable to verify whether all putative TfpM enzymes were active and capable of transferring the pneumococcal CPS8 like TfpMMo. Previous studies with further-truncated EPA-ComP constructs showed that they were more stable than the fusion protein that only lacked the N-terminal pilin membrane domain (Knoot et al. 2021). It is possible further truncation could also be used to stabilize the TfpM-associated pilins. Given the similar genetic architecture of the pilin-TfpM pairs and similar size of the TfpM proteins to TfpO protein, we initially hypothesized that TfpM proteins would only transfer short oligosaccharides like TfpO proteins. Much to our surprise, we observed that TfpM proteins can transfer long-chain polysaccharides (Fig. 4G). TfpO proteins are limited to transferring oligosaccharides between 3 and 6 sugars, depending on the monosaccharide composition (Castric et al. 2001; Comer Jason et al. 2002; Faridmoayer et al. 2007; Harding et al. 2015). In contrast, our western blot data demonstrate that TfpMMo can transfer glycans ranging from one to greater than 30 CPS8 tetrasaccharide repeat units. Thus, TfpM proteins share qualities of both TfpO and PglS/PglL OTases: TfpM proteins are similar in size to TfpO and glycosylate a C-terminal hydroxy-amino acid (serine or threonine) yet, like PglL and PglS, will transfer long polysaccharide chains to acceptor proteins (Table 2). The ability of TfpM to transfer long-chain polysaccharide shows that this ability is not limited to the PglS and PglL OTase families.
Table 2.
General characteristics of bacterial O-linking OTases.
| OTase | Protein size (amino acids) | Sugar monomers transferred | Glycan reducing-end tolerance | Native substrate protein(s) | Glycosylation site | References |
|---|---|---|---|---|---|---|
| TfpM | 420–440 | No known limits | Glc, Gal, 2-N-acetyl sugars | Type IV pilin-like proteins | C-terminal serine or threonine | This work |
| TfpO (PilO) | 440–470 | Roughly 3–6 | 2-N-acetyl sugars, possibly more | PilA | C-terminal serine or threonine | Castric et al. (2001), Comer Jason et al. (2002), DiGiandomenico et al. (2002), Nguyen et al. (2012), Harding et al. (2015) |
| PglL | 550–600 | No known limits | Gal, 2-N-acetyl sugars | Many, including PilE | Protein-internal sequon | Faridmoayer et al. (2007), Faridmoayer et al. (2008), Vik et al. (2009), Hayes et al. (2021) |
| PglS | 520–550 | No known limits | Glc, Gal, 2-N-acetyl sugars | ComP | Protein-internal sequon | Harding et al. (2015), Harding et al. (2019) |
Our murine vaccination studies show that a TfpMMo-derived bioconjugate elicits a GBSIII IgG response. Thus far, bioconjugates have only been produced using sequons that reside in the interior of carrier proteins, but our data show that C-terminally glycosylated carriers could also be used to produce bioconjugates that elicit an immune response. Furthermore, the availability of 2 different types of O-linking OTases, i.e. PglL/PglS and TfpM, for glycoconjugate vaccine production could increase the versatility of the process, for example in cases where one OTase enzyme is unable to transfer a specific glycan but the other is. An extension of this approach would be to use 2 different OTases in one expression system, as we have shown in this work.
It remains an outstanding question as to why TfpO proteins, but not TfpM proteins, are limited to transferring only small oligosaccharides to acceptor proteins. An alignment of TfpMMo and TfpO from Acinetobacter nosocomialis M2 (TfpOM2) shows that both enzymes are of similar size and are characterized by a wzy_C superfamily protein domain roughly spanning residues 240–320 (Supplementary Fig. 3). Aside from a small amino acid extension present at the C-terminus of TfpOM2, there are no immediately obvious differences in the primary structure to explain the different glycan size transfer capabilities. Interestingly, a comparison of in silico models of TfpMMo and TfpOM2 generated using RoseTTAFold (Baek et al. 2021) suggest a possible structural explanation for the different glycan-transfer abilities. In the resulting RoseTTAFold models, identification of the putative active site and lipid-linked oligosaccharide binding site of the 2 OTases is possible by comparing them to the recently published structure of a bacterial WaaL enzyme, which also contains a wzy_C domain (Ashraf et al. 2022). The resulting RoseTTAfold models of TfpOM2 contain a structural “plug”-like feature present near the active site of TfpOM2 that is absent in the TfpMMo models (Supplementary Fig. 9). We hypothesize that the presence of this “plug”-like structure in TfpO enzymes, but not TfpM enzymes, may hinder or block the binding of longer lipid-linked glycans in the active site of the OTase. Further experiments are required to test this hypothesis.
Studies have shown that a minimum number of monosaccharides must be present in the glycoconjugate, typically 10 or more, for proper docking of the glycan to the B-cell receptors (Anish et al. 2021). The size of bacterial O-antigen and capsule glycan repeat units varies but they typically contain 3–6 monosaccharides (Liu et al. 2020). Our murine vaccination studies show that a TfpMMo-derived bioconjugate elicits a GBSIII IgG response (Fig. 6). Thus far, bioconjugates have only been produced using sequons that reside in the interior of carrier proteins, but our data suggest that C-terminally glycosylated carriers could also be used to produce bioconjugates that elicit an immune response. The ability to transfer multi-repeat unit glycans could allow TfpM proteins, but likely not TfpO proteins, to be used to produce glycoconjugate vaccines. Furthermore, the availability of 2 different types of O-linking OTases, i.e. PglL/PglS and TfpM, for glycoconjugate vaccine production could increase the versatility of the process, for example in cases where 1 OTase enzyme is unable to transfer a specific glycan, but the other is. An extension of this approach would be to use 2 different OTases in one expression system, as we have shown in this work.
The protein mechanisms or structural features that determine whether a given OTase can transfer certain glycans, but not others, is not well understood. Furthermore, whether glycan substrate specificity is related to the number of repeat units capable of being transferred by an OTase is also poorly understood. We show that TfpMMo is like PglSADP1 in its ability to transfer diverse glycans with different reducing-end sugars, repeat unit structures, and branched or unbranched polymers. Interestingly, PglSADP1 and PglLNm are both much larger proteins than TfpMMo, namely 548 and 620 vs. 425 amino acids, respectively. Sequence alignments comparing the transmembrane and solvent-exposed domains of PglSADP1 to TfpMMo (Dobson et al. 2015) show that PglSADP1 has a larger predicted C-terminal periplasmic domain that is absent in TfpMMo. Since both proteins can perform similar functions, this region may have additional roles in PglSADP1, possibly being involved in recognizing protein sequons in the interior of substrates. Unlike PglSADP1, TfpMMo recognizes the C-terminal tail of its substrate pilin, which may rely on a different structural motif for protein recognition and glycosylation specificity.
The sequential truncation of the PilMo provides several insights into this protein and orthologs from other TfpM-encoding strains. Our experiments demonstrate that PilMo can be truncated to 20 amino acids while retaining glycosylation levels on par with that of the longer PilMo fragment. Pil20 contains the DSL region that, from sequence alignments, is also present in several other types of pilins, including Acinetobacter soli CIP110264 ComP and P. aeruginosa 1244 PilA (Supplementary Fig. 8). Although smaller fragments of PilMo were able to be glycosylated by TfpMMo, the levels were below those of the PilMoΔ28 or Pil20 variants suggesting a preference for the intact DSL region. The ability of TfpMMo to glycosylate Pil3 but not Pil2 is in line with previous studies of P. aeruginosa 1244 PilA where a 3-amino acid fragment was able to be glycosylated when fused to C-terminus of another pilin protein (Harvey et al. 2009). The inability of TfpMMo to glycosylate Pil2 could be due to myriad reasons. PilA glycosylation by TfpO was previously shown to require a minimum number of amino acids between the second cysteine in the disulfide and C-terminus (Horzempa et al. 2006). It was hypothesized that placing the serine too close to the rest of the pilin structure results in a steric hindrance that blocks TfpO glycosylation (Horzempa et al. 2006). A similar effect could be occurring in Pil2. Notably, the residue that is deleted from Pil3 to generate Pil2 is an arginine, R165 (Fig. 5A). The C-terminal residue of EPA is a lysine residue, which one might expect could substitute for R165 deletion in the conversion of Pil3 to Pil2 (Supplementary Fig. 8A). However, this is not to be the case suggesting that the interaction between TfpMMo and PilMo is finely tuned.
In conclusion, we have discovered a novel family of bacterial O-linking OTases that share features of both TfpO proteins and PglS and PglL proteins. The identification of TfpM proteins opens inroads to dissect the mechanisms of substrate specificity in bacterial O-linking OTases, both in terms of what limits glycan length and how these enzymes recognize the reducing end of glycan substrates. The results herein show that the ability to transfer long glycan substrates to acceptor proteins is not limited to PglS and PglL proteins. Indeed, TfpO proteins are thus far the only O-linking OTases of the 4 known families limited to transferring glycans with low numbers of repeat units. TfpO and TpfM proteins have moderate sequence similarity, similar size, and recognize C-terminal sequons on substrate proteins. The major known differences are the ability of TfpM proteins to transfer longer glycans as well as transfer virtually any glycan, including those with glucose at the reducing end. As such, comparative studies of the TfpM and TfpO enzyme families could provide important insights into the features that regulate glycan substrate size. Finally, TfpMMo has a similar broad substrate scope to PglSADP1 suggesting that this enzyme could be leveraged as a powerful tool for protein glycoengineering.
Materials and methods
Cloning and plasmid assembly
All primers and oligos used in this study are listed in Supplementary Table 1. Working antibiotic concentrations used for liquid culture and in Luria broth agar (LB-agar) plates were as follows: ampicillin (Amp), 100 μg/mL, kanamycin (Kan), 20 μg/mL, tetracycline (Tet), 10 μg/mL, spectinomycin (Sp), 50 μg/mL. For cloning the tfpM pilin-OTase genes, we ordered HiFi gblocks (Integrated DNA Technologies, IDT) designed with terminal 25 base pair overlaps for Gibson assembly with PCR-linearized plasmid. The plasmid backbone for these fragments was amplified from a pEXT20 plasmid (Dykxhoorn et al. 1996) encoding the P. aeruginosa EPA gene under control of a tac promoter (pVNM57) (Knoot et al. 2021). The EPA gene has a deletion of residue E553 resulting in an inactivated toxin. The linearized plasmid was mixed separately with each of the synthesized tfpM gBlocks and assembled using an NEBuilder HiFi DNA Assembly Kit (New England Biolabs, NEB). After assembly, the plasmids were transformed into E. coli Stellar cells (Takara Bio) by heat shock, out-grown for an hour at 37 °C, and plated on LB-agar supplemented with Amp. Individual colonies were picked and grown in LB media with appropriate antibiotic and plasmids isolated using a GeneJet Plasmid Miniprep Kit (Thermo Fisher). All plasmids were sequence-verified by Sanger sequencing (Genewiz). The plasmid expressing the M. osloensis 1202 EPA-Pil∆28 fusion and TfpMMo was named pVNM227. To generate the PilMo site-directed mutants, we designed overlapping PCR primers that introduced the necessary codon changes in the pilin gene and amplified each fragment from pVNM227 plasmid. The resulting PCR product was DpnI-digested (NEB) for 30 min at 37 °C and gel-purified from agarose gel using a Pure-Link Gel Extraction Kit (Thermo Fisher). To insert the truncated pilin gene regions, we ordered complementary oligos with terminal 25 bp overlaps homologous to the pVNM227 PCR product. The oligos were re-suspended in purified water, mixed, and annealed together in thermocycler by heating to 98 °C for 5 min followed by a slow cooling to 4 °C at 0.1 °C/min. The annealed oligos were diluted 1–5 in water and assembled with PCR-linearized pVNM227 using an NEBuilder HiFi DNA Assembly Kit (NEB). The resulting DNA was transformed into Stellar cells and plasmids isolated and verified as described above. The plasmid encoding EPA-Pil20 and TfpM was termed pVNM297. The N-terminally His-tagged EPA-Pil20 variant was constructed by linearizing pVNM297 using PCR and using this fragment in Gibson assembly with complementary annealed oligos containing the 6xHis coding region and terminal homologous regions, resulting in pVNM291. pVNM167 was generated by digesting the previously described EPAiGTcc plasmid (Knoot et al. 2021) with SalI. The purified SalI fragment was Gibson assembled with the pglS gene with its native 100 bp 5′ UTR amplified from A. baylyi ADP1 gDNA. pVNM245 was generated from pVNM167 template by separate PCR reactions to amplify products with overhangs for Gibson assembly: (i) the vector backbone with PglS and EPA with one iGT, (ii) the second iGT for integration between E548 and G549, and (iii) the C-terminus of EPA downstream of the iGT. The plasmid pVNM337 was created by amplifying tfpM from pVNM291 with the primers EPA 3′ F1 and pglS-tfpM R1 and cloning the product into PCR-linearized pVNM167, which was amplified with pglS 5′ F1 and EPA 3′ R1. Phylogenetic trees for the TfpM and pilin proteins were generated using the phylogeny.fr server (http://www.phylogeny.fr/), which uses MUSCLE, PhyML, and TreeDyn for sequence alignment, tree calculation, and image generation, respectively.
Expression of glycans and cloning of K. pneumoniae O2a glycan genes
The S. pneumonia CPS8 glycan was expressed from plasmid pB8 (TetR) (Kay et al. 2016), the S. enterica LT2 glycan from plasmid pPR1347 (KanR) (Neal et al. 1993), the E. coli O16 wbbL gene from plasmid pMF19 (SpR) (Feldman et al. 2005), and the GBSIII glycan was from a pBBR1MCS2 derivative we reported recently (Duke et al. 2021). Bioconjugation with the K. pneumoniae O2a O-antigen has not previously been reported. To clone the genes that encode the machinery required to synthesize the O2a glycan, we PCR-amplified the wzm, wzt, wbbM, glf, wbbN, and wbbO genes (Clarke et al. 2018) from K. pneumoniae strain NTUH K2044 genomic DNA. K. pneumoniae was cultured to saturation in LB media overnight and genomic DNA was isolated using a Wizard Genomic DNA Purification Kit (Promega). The plasmid backbone for the O2a cluster was amplified from plasmid pBBR1MCS2 (KanR) (Kovach et al. 1995). Primers for these reactions are listed in Supplementary Table 1. The PCR products from these reactions were assembled using Gibson assembly with a NEBuilder HiFi DNA Assembly Kit (NEB). Stellar cells were transformed and plasmid isolated and verified as described in the previous section.
Bioconjugation and western blots
E. coli strains used the bioconjugation experiments were either SDB1 or CLM24 (Feldman et al. 2005). SDB1 is a W3110 E. coli derivative with mutations in the genes encoding for WecA, the glycosyltransferase that initiates synthesis of the endogenous E. coli O16 antigen and WaaL, the enzyme transferring Und-PP linked glycans to Lipid A-core saccharide to produce LPS. CLM24 is a W3110 derivative with only a deletion of waaL. Elimination of these genes prevents crosstalk of the heterologous bioconjugation system and endogenous E. coli glycosylation pathways. To prepare E. coli strains for bioconjugation, we electroporated plasmids using competent cells prepared as previously described (Knoot et al. 2021) followed by out-growth at 37 °C in SOB media. The cells were plated on LB-agar with appropriate antibiotics. The next day, 8–10 colonies were picked and inoculated into LB or TB with antibiotics and grown overnight while shaking at 30 °C. The next morning, starter cultures were inoculated into either 30 mL media in a 125 mL Erlenmeyer flask or 1 L media in a 2 L flask to a starting optical density at 600 nm (OD600) of 0.05. Cultures were grown while shaking at 175 RPM until the OD600 reached 0.4–0.6 at which point the cultures were induced with 1 mM IPTG. All bioconjugation experiments were performed at 30 °C unless otherwise noted. After overnight induction, amounting to 20–24 h total growth, the OD600 was measured, and 0.5 OD units of cells pelleted for analysis.
The cell pellets were suspended in 100 μL 1× Laemmli Buffer (Biorad) and boiled for 10 min at 100 °C. The boiled samples were briefly centrifuged at 10,000 rcf and equivalent amounts, normalized to the same OD600 per lane, loaded for SDS-PAGE separation on a 7.5% Mini-Protean TGX gel (Biorad). Proteins were transferred to a nitrocellulose membrane using a semi-dry electrode system and blocked with Intercept Blocking Buffer (Li-Cor) for 1 h. The membrane was incubated with primary antibodies in 1:1 blocking and TBST for 45 min. For protein detection, we used commercial rabbit anti-EPA and mouse anti-6xHis antibodies (Millipore-Sigma). Rabbit glycan antibodies for CPS8, GBSIII, and O16 were purchased from SSI Diagnostica. K. pneumoniae rabbit O2a antibodies were a generous gift from Prof. Chris Whitfield (University of Guelph) (Clarke et al. 2018). Salmonella Group B rabbit antibodies were purchased from BD. Primary incubation was followed by 3 washes with TBST buffer totaling 15 min The membranes were then incubated with secondary antibodies IRDye 680RD goat anti-mouse and/or IRDye 800CW goat anti-rabbit (Li-Cor) in 1:1 blocking buffer and TBST for 30 min. After a final 15-min TBST wash, the membranes were imaged using a Li-Cor Odyssey CLx.
Lys-C digestion of recombinant M. osloensis PilMo∆28
In-gel digestion was accomplished according to the protocol of Shevchenko et al. (2006) with minor modifications. Gel-separated glycosylated EPA-Pil∆28-CPS8 was excised and destained with destaining solution (50 mM NH4HCO3, 50% ethanol) twice for 10 min at room temperature with shaking at 750 RPM. The destained band was then dehydrated with 100% ethanol for 10 min and dried by vacuum-centrifugation for 10 min before being rehydrated in 10 mM DTT in 50 mM NH4HCO3. Reduction was carried out for 60 min at 56 °C after which the gel band was dehydrated twice with 100% ethanol for 10 min to remove the remaining reduction buffer. The reduced sample was then sequentially alkylated with 55 mM iodoacetamide in 50 mM NH4HCO3 for 45 min at RT in the dark. The alkylated sample was then washed 4 times for 10 min with 50 mM NH4HCO3 followed by 100% ethanol, followed by 50 mM NH4HCO3 followed by 100% ethanol before being dried by vacuum-centrifugation. The dried alkylated sample was then rehydrated with 20 ng/μL Lys-C endoprotease (Wako Chemicals) in 40 mM NH4HCO3 at 4 °C for 1 h. Excess Lys-C was removed, gel pieces were covered in 40 mM NH4HCO3 and incubated overnight at 37 °C. Peptides were concentrated and desalted using C18 stage tips (Ishihama et al. 2006; Rappsilber et al. 2007) then eluted in buffer B (0.5% acetic acid, 80% acetonitrile (ACN)) before being dried and stored at −20 °C prior to LC–MS analysis.
Analysis of recombinant M. osloensis PilMo∆28 using reversed phase LC–MS
The C18-concentrated digest was resuspended in buffer A* (0.1% TFA, 2% ACN) and separated using a 2-column chromatography set up comprising a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). The sample was concentrated onto the trap column at 5 μL/min using 0.1% formic acid (FA) for 5 min and infused into an Orbitrap Fusion Lumos Tribrid Mass Spectrometer equipped with a FAIMS Pro interface (Thermo Fisher Scientific) at 300 nL/min via the analytical column using a Dionex Ultimate 3000 UPLC (Thermo Fisher Scientific) by altering the concentration of buffer A (2% DMSO, 0.1% FA) and buffer B (78% ACN, 2% DMSO and 0.1% FA). Identification of potential glycopeptides utilized a 140-min analytical run while targeted analysis utilized a 60-min run. Within the identification analytical run the buffer composition was altered from 3% buffer B to 28% buffer B over 120 min, 28% buffer B to 40% buffer B over 9 min, 40% buffer B to 100% buffer B over 3 min, then the composition was held at 100% buffer B for 2 min, and then dropped to 3% buffer B over 2 min and held at 3% buffer B for another 8 min The Lumos Mass Spectrometer was operated in a stepped FAIMS data-dependent mode at 3 different FAIMS CVs, −25, −45, and −65 as previously described (Ahmad Izaham et al. 2021), switching between the acquisition of a single Orbitrap MS scan (60 k resolution) every 1.5 s followed by Orbitrap HCD scans (maximum fill time 120 ms, AGC 2 × 105 with a resolution of 30 k for Orbitrap MS–MS scans and an NCE of 25, 30, 45) at each of the 3 FAIMS CVs. For targeted characterization of PilMo glycopeptides an analytical run altering the buffer composition from 3% buffer B to 15% buffer B over 30 min, 15% buffer B to 30% buffer B over 10 min, 30% buffer B to 80% buffer B over 5 min, then the composition was held at 100% buffer B for 5 min, and then dropped to 3% buffer B over 1 min and held at 3% buffer B for another 9 min was undertaken. Parallel reaction monitoring using HCD (maximum fill time 250 ms, AGC 2.5 × 105 with a resolution of 30 k for the Orbitrap MS–MS scan and an NCE of 15, 30, 35) and EThcD (maximum fill time 250 ms, AGC 2.5 × 105 with a resolution of 30 k for the Orbitrap MS–MS scan and Electron-transfer dissociation (ETD) reaction times controlled using calibration charge dependent ETD parameters (Rose et al. 2015) of the +2 charge state of the HexHexA modified glycopeptide 762FLPANCRGT770 (687.2972 m/z) was undertaken with a FAIMS CV of −45.
Open searching of PilMo∆28 and the annotation of HexHAxA-modified FLPANCRGT
The identification of PilMo glycosylation events was accomplished using open database searching as previously described (Lewis et al. 2021). Briefly, datafiles were processed with MSfragger 3.4 (Kong et al. 2017; Polasky et al. 2020) in FragPipe (version 17.1) searching against the M. osloensis PilMo sequence (NCBI Accession: WP_156627541.1). Searches were undertaken using “Lys-C” enzyme specificity with carbamidomethylation of cysteine as a fixed modification and oxidation of methionine as a variable modification with a maximum of 2 missed cleavages allowed. To enable identification of potential glycosylation events a mass tolerance, referred to as a delta mass, of 0–2,000 Da was allowed. Delta masses observed on the C-terminal peptide 762FLPANCRGT770 were manually inspected to identify potential glycosylation events. Parallel reaction monitoring results corresponding to the HexHexA-modified glycopeptide 762FLPANCRGT770 was manually extracted using the Freestyle Viewer (1.7 SP1, Thermo Fisher Scientific), the MS/MS data annotated using the Interactive Peptide Spectral Annotator (Brademan et al. 2019) (http://www.interactivepeptidespectralannotator.com/PeptideAnnotator.html). Annotation of spectra allowed for the modification of the terminal T residue with HexHexA (338.0849 Da) as well as Hex (162.0528 Da). The resulting MS data and search results have been deposited into the PRIDE ProteomeXchange Consortium repository (Perez-Riverol et al. 2015, 2019) and can be accessed with the identifier PXD033468.
M. osloensis CCUG 350 glycoproteomic sample preparation
M. osloensis CCUG 350 (ATCC 19976) was obtained from the American Type Culture Collection (ATCC 19976) and cultured in Bovine Heart Infusion media (BD Difco) at 37 °C while shaking at 75 RPM. The cultures were grown for 6 days then collected by centrifugation at 1,000 rcf for 5 min. The cell pellet was washed in cold PBS 3 times then suspended in SDS lysis buffer (4% SDS, 0.1 M Tris–HCl pH 8.5, 10 mM DTT) and boiled 5 min with intermittent vortexing. 500 μg of protein from this lysate was precipitated using 2 washes with ice-cold acetone and intervening overnight storage at −20 °C. The acetone-precipitated M. osloensis CCUG 350 samples were processed using the in-StageTip preparation approach as previously described (Kulak et al. 2014). Cells were resuspended in 4% sodium deoxycholate (SDC), 100 mM Tris pH 8.0 and boiled at 95 °C with shaking for 10 min to solubilize the proteome. Samples were allowed to cool for 10 min and then boiled for a further 10 min before the protein concentration was determined by bicinchoninic acid assays (Thermo Fisher Scientific). Samples were reduced/ alkylated with the addition of Tris-2-carboxyethyl phosphine hydrochloride and Chloroacetamide (final concentration 10 and 40 mM, respectively), and samples incubated in the dark for 1 h at 45 °C. Following reduction/alkylation samples were digested overnight with Lys-C (1/50 w/w Wako chemicals) at 37 °C with shaking at 1,000 rpm. Digested samples were then mixed with 1.25 volumes of isopropanol to prevent the precipitation of SDC (Humphrey et al. 2018) and samples acidified with 10% TFA to a final concentration of 1% TFA. Home-made SDB-RPS StageTips were prepared according to previously described protocols (Rappsilber et al. 2007; Kulak et al. 2014; Harney et al. 2019) with 6 discs of SDB-RPS excised using a blunt 16-gauge Hamilton needle and logged in a 200 μL tip. SDB-RPS StageTips were placed in a Spin96 tip holder (Harney et al. 2019) to enable batch-based spinning of samples and tips conditioned with 100% acetonitrile; followed by 30% methanol, 1% TFA followed by 90% isopropanol, 1% TFA with each wash spun through the column at 1,000 × g for 3 min. Acidified isopropanol/peptide mixtures were loaded onto the SDB-RPS columns and spun through the tips washed with 90% isopropanol, 1% TFA followed by 1% TFA in Milli-Q water. Peptides were eluted with 80% acetonitrile, 5% ammonium hydroxide and dried by vacuum centrifugation at room temperature before being stored at −20 °C.
FAIMS-based glycoproteomic analysis
Glycosylation analysis of M. osloensis CCUG 350 was undertaken with FAIMS-based fractionation as previously described (Ahmad Izaham et al. 2021). Briefly M. osloensis proteome samples were resuspended in Buffer A* (2% acetonitrile, 0.1% TFA) and 2 μg of peptide used for each FAIMS CV. Peptide samples were separated using a 2-column chromatography set up composed of a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μL/min for 5 min with Buffer A (0.1% formic acid, 2% DMSO) and then infused into an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific) equipped with a FAIMS Pro interface at 300 nL/min via the analytical column using a Dionex Ultimate 3000 UPLC (Thermo Fisher Scientific). 125-min analytical runs were undertaken by altering the buffer composition from 2% Buffer B (0.1% formic acid, 77.9% acetonitrile, 2% DMSO) to 23% B over 95 min, then from 23% B to 40% B over 10 min, then from 40% B to 80% B over 7 min. The composition was held at 80% B for 3 min, and then dropped to 2% B over 1 min before being held at 2% B for another 9 min. The Lumos mass spectrometer was operated in a static FAIMS data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan (120 k resolution) every 3 s and HCD MS2 events (FTMS, 30 k resolution, maximum fill time 80 ms, normalize collision energy 30, AGC of 250%). HCD scans observed to contain oxonium ions (204.0867; 138.0545; and 366.1396 m/z) or the predicted y7+2 ion (388.1794 m/z) of the C-terminal pilin peptide 762FLPANCRGT770 triggered product-dependent MS/MS analysis (Saba et al. 2012) with 3 additional scans; a Orbitrap EThcD scan (NCE 15%, maximal injection time of 250 ms, AGC 2*105 with a resolution of 30 k with the extended mass range setting enabled to improve the detection of high mass glycopeptide fragment ions) (Čaval et al. 2019); a ion trap CID scan (NCE 35%, maximal injection time of 40 ms, AGC 5*104) and a stepped collision energy HCD scan (using NCE 35% with 8% Stepping, maximal injection time of 250 ms, AGC 2*105 with a resolution of 30 k). A total of 6 FAIMS CVs were acquired; −20, −30, −40, −50, −60, and −70. To enable the characterization of potential pilin glycopeptides parallel reaction monitoring was undertaken with a FAIMS CV of −30 using HCD, (maximum fill time 300 ms, AGC 2.5 × 105 with a resolution of 50 k for the Orbitrap MS–MS scan and an NCE of 15, 30, 35); EThcD (maximum fill time 300 ms, AGC 2.5 × 105 with a resolution of 50 k for the Orbitrap MS–MS scan with 150 ms ETD reaction times) or CID (maximum fill time 300 ms, AGC 2.5 × 105 with a resolution of 50 k for the Orbitrap MS–MS scan). Summed PRM scans corresponding to the ions 919.9163 and 1000.9434 m/z were manually extracted using the Freestyle Viewer (1.7 SP1, Thermo Fisher Scientific) and the MS/MS data annotated using the Interactive Peptide Spectral Annotator (Brademan et al. 2019) (http://www.interactivepeptidespectralannotator.com/PeptideAnnotator.html). CID spectra were manually annotated to assign glycan compositions for the m/z 939.9195; 919.9163; and 1000.9434. Open searching of FAIMS fractions was also undertaken using MSfragger as above against the M. osloensis ATCC 19976 proteome (Uniprot: UP000255230). The resulting MS data and search results have been deposited into the PRIDE ProteomeXchange Consortium repository (Perez-Riverol et al. 2015, 2019) and can be accessed with the identifier PXD033468.
Bioconjugate protein purification
Cells for protein purification were grown in 1 L TB media and bioconjugates isolated using an osmotic shock protocol. After overnight growth and induction, the cells were pelleted by centrifugation, and washed in 0.9% NaCl. The washed cell pellets were suspended in 200 mM Tris–HCl pH 8.5, 100 mM EDTA, 25% sucrose and incubated while rolling for 30 min at 4 °C. Cells were pelleted by centrifugation at 4,700 rcf for 30 min and the resulting pellet suspended in 20 mM Tris–HCl pH 8.5 and incubated while rolling for 45 min at 4 °C. The suspension was centrifuged for 30 min at 18,000 rcf. The supernatant containing the periplasmic fraction was concentrated and either loaded directly on an FPLC anion-exchange column or, for His-tagged EPA-Pil∆28 bioconjugates, purified using Nickel IMAC as previously described (Knoot et al. 2021). The periplasmic extract or IMAC eluate was concentrated and buffer-exchanged into 20 mM Tris–HCl pH 8.0, filtered through a 0.2 μm PES filter then loaded on an Äkta pure FPLC instrument (Cytiva) equipped with a SOURCE 15Q 4.6/100 PE anion-exchange column (Cytiva). The bioconjugates were eluted at 2 mL/min using a stepwise gradient with buffer A (20 mM Tris pH 8) and buffer B (20 mM Tris pH 8, 1 M NaCl) from 0% B to 25% in 5% increments at 10 column volumes for each concentration. Bioconjugates for immunization were further purified using a Superdex 200 Increase 10/300 GL column. The concentrated bioconjugates pooled from the anion-exchange column were loaded on a pre-equilibrated Superdex 200 column in PBS buffer and eluted at a flow rate of 0.75 mL/min Fractions containing the purified bioconjugates were pooled, concentrated, and frozen at −80 °C for storage. Protein concentrations for immunization and western blots were determined using a Pierce BCA Protein Assay kit (Thermo Fisher). The ratio of polysaccharide to protein calculate for vaccine dosing was determined using the method described in Duke et al. (2021).
Murine immunization
All murine immunizations followed ethical regulations for animal testing and research. Experiments were carried out at Washington University School of Medicine in St. Louis according to the institutional guidelines and received approval from the Institutional Animal Care and Use Committee at Washington University in St. Louis. Five-week-old female CD-1 outbred mice (Charles River Laboratories) were subcutaneously injected with 100 μL of a vaccine formulation on days 0, 14, and 28. The vaccination groups were EPA-Pil20 alone (5 μg protein) and EPA-Pil20-GBSIII (5 μg protein, 1 μg polysaccharide). Mice had sera collected on days 0, 14, 28, and 42. All vaccines were formulated with Alhydrogel 2% aluminum hydroxide gel (InvivoGen) at a 1:9 ratio (50 μL vaccine to 5.5 μL alum in 44.5 μL 1× sterile phosphate buffered saline).
Enzyme-linked immunosorbent assays
IgG kinetic titers were determined using enzyme-linked immunosorbent assay (ELISA). Briefly, 96-well plates (TRP Immunomaxi plates) were coated in triplicate overnight with approximately 106 CFU/100 μL of glycoengineered SDB1 E. coli expressing the GBSIII capsular polysaccharide in sodium carbonate buffer. The coating E. coli strain was grown the same as referenced above and after overnight 1 mM IPTG induction to induce GBSIII expression was washed and diluted to coat plates. Wells were blocked with 1% BSA in PBS and washed with 0.05% PBS-Tween (PBST), all subsequent washes were the same. Serum from mice was diluted to 1:100 and added to wells for 1 h at room temperature then washed. Total IgG titers were detected by HRP conjugated anti-mouse IgG (GE Lifesciences, 1:5,000 dilution) added to wells for 1 h at room temperature. After washing, plates were developed using 3,3′,5,5′ tetramethyl benzidine (TMB) substrate (Biolegend) and stopped with 2 N H2SO4. The optical densities were determined at 450 nm using a microplate reader (Bio-Tek). Total IgG product was determined using IgG standards to generate a standard curve for data fitting. Standard wells were coated with IgG in sodium carbonate buffer and treated the same as sample wells thereafter. All wells were normalized to blank wells that were treated the same as all samples wells minus receiving the primary mouse sera. Significance was determined using Mann–Whitney nonparametric test with P < 0.05.
In silico modelling of TfpMMo and TfpOM2 using RoseTTAFold
Full-length amino acid sequences of A. nosocomialis M2 TfpO (WP_022575995) and M. osloensis TfpMMo (WP_156627541) were submitted to the Robetta server (https://robetta.bakerlab.org/submit.php) and modelled using RoseTTAFold (Baek et al. 2021). The resulting 3D models of each protein were aligned in Pymol 2.5.2 using the super console command (https://pymol.org/2/).
Data availability statement
The data underlying this article are available in the article and in its online supplementary material.
Abbreviations
OTase, oligosaccharyltransferase; EPA, exotoxin A from P. aeruginosa; CPS8, S. pneumoniae capsular polysaccharide 8; GBSIII, Group B Streptococcus capsular polysaccharide III; LT2, S. enterica Group B serotype LT2 O-antigen; O16, Escherichia coli serotype O16 O-antigen; O2a, K. pneumoniae serotype O2a O-antigen; Glc, glucose; Gal, galactose; Rha, rhamnose; GlcNAc, N-acetylglucosamine; Abe, abequose; NeuNAc, N-acetylneuraminic acid, sialic acid; DSL, disulfide loop (present near the C-terminus of type IV pilins); HCD, higher-energy C-trap dissociation; EThcD, electron-transfer/higher-energy collision dissociation; FAIMS, high-field asymmetric waveform ion mobility spectrometry.
Accession numbers
NCBI protein accession numbers for other proteins discussed in this study: A. baylyi ADP1 PglS, WP_004923783; A. baylyi ADP1 PglL, WP_004930753; Neisseria meningitidis MC58 PglL, WP_002244038; A. nosocomialis M2 TfpO, WP_022575995; P. aeruginosa 1244 TfpO, WP_058150674; A. soli CIP110264 ComP, WP_004934263; N. meningitidis M2 PilA, WP_022575996; P. aeruginosa 1244 PilA, CAA58768.
Supplementary Material
Acknowledgements
We thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute for access to MS instrumentation.
Contributor Information
Cory J Knoot, Omniose, 4340 Duncan Ave, Suite 202, St. Louis, MO 63110, USA.
Paeton L Wantuch, Department of Pediatrics, Division of Infectious Diseases, Washington University School of Medicine, 4990 Children’s Place, St. Louis, MO 63110, USA.
Lloyd S Robinson, Omniose, 4340 Duncan Ave, Suite 202, St. Louis, MO 63110, USA.
David A Rosen, Department of Pediatrics, Division of Infectious Diseases, Washington University School of Medicine, 4990 Children’s Place, St. Louis, MO 63110, USA; Department of Molecular Microbiology, Washington University School of Medicine, 660 Euclid Ave, St. Louis, MO 63110, USA.
Nichollas E Scott, Department of Microbiology and Immunology, University of Melbourne at the Peter Doherty Institute for Infection and Immunity, Parkville, VIC 3010, Australia.
Christian M Harding, Omniose, 4340 Duncan Ave, Suite 202, St. Louis, MO 63110, USA.
Funding
This work was funded by National Institute of Allergy and Infectious Disease SBIR project number R44AI131742 awarded to VaxNewMo, LLC. NES is supported by an Australian Research Council Future Fellowship (FT200100270) and an ARC Discovery Project Grant (DP210100362). PLW is a W.M. Keck Fellow at the Washington University School of Medicine in St. Louis.
Conflict of interest statement
CJK, LSR, and CMH have a financial stake in Omniose, a for-profit entity developing bioconjugate vaccines using patented technology derived from the data presented in this and other published manuscripts.
References
- Ahmad Izaham AR, Ang CS, Nie S, Bird LE, Williamson NA, Scott NE. What are we missing by using hydrophilic enrichment? Improving bacterial glycoproteome coverage using total proteome and FAIMS analyses. J Proteome Res. 2021:20:599–612. [DOI] [PubMed] [Google Scholar]
- Anish C, Beurret M, Poolman J. Combined effects of glycan chain length and linkage type on the immunogenicity of glycoconjugate vaccines. NPJ Vaccines. 2021:6:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K et al. Structural basis of lipopolysaccharide maturation by the O-antigen ligase. Nature. 2022:604:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee Gyu R, Wang J, Cong Q, Kinch Lisa N, Schaeffer RD et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021:373:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brademan DR, Riley NM, Kwiecien NW, Coon JJ. Interactive peptide spectral annotator: a versatile web-based tool for proteomic applications. Mol Cell Proteomics. 2019:18:S193–S201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric P. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 1995:141:1247–1254. [DOI] [PubMed] [Google Scholar]
- Castric P, Cassels FJ, Carlson RW. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J Biol Chem. 2001:276:26479–26485. [DOI] [PubMed] [Google Scholar]
- Čaval T, Zhu J, Heck AJR. Simply extending the mass range in electron transfer higher energy collisional dissociation increases confidence in N-glycopeptide identification. Anal Chem. 2019:91:10401–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick JM, Kolippakkam D, Nusinow DP, Zhai B, Rad R, Huttlin EL, Gygi SP. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat Biotechnol. 2015:33:743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Ovchinnikova OG, Kelly SD, Williamson ML, Butler JE, Liu B, Wang L, Gou X, Follador R, Lowary TL et al. Molecular basis for the structural diversity in serogroup O2-antigen polysaccharides in Klebsiella pneumoniae. J Biol Chem. 2018:293:4666–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer Jason E, Marshall Mark A, Blanch Vincent J, Deal Carolyn D, Castric P. Identification of the Pseudomonas aeruginosa 1244 pilin glycosylation site. Infect Immun. 2002:70:2837–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curd H, Liu D, Reeves PR. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998:180:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiandomenico A, Matewish MJ, Bisaillon A, Stehle JR, Lam JS, Castric P. Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol Microbiol. 2002:46:519–530. [DOI] [PubMed] [Google Scholar]
- Dobson L, Reményi I, Tusnády GE. CCTOP: a consensus constrained topology prediction web server. Nucleic Acids Res. 2015:43:W408–W412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke JA, Paschall AV, Robinson LS, Knoot CJ, Vinogradov E, Scott NE, Feldman MF, Avci FY, Harding CM. Development and immunogenicity of a prototype multivalent group B Streptococcus bioconjugate vaccine. ACS Infectious Diseases. 2021:7:3111–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, St. Pierre R, Linn T. A set of compatible tac promoter expression vectors. Gene. 1996:177:133–136. [DOI] [PubMed] [Google Scholar]
- Faridmoayer A, Fentabil Messele A, Mills Dominic C, Klassen John S, Feldman MF. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol. 2007:189:8088–8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridmoayer A, Fentabil MA, Haurat MF, Yi W, Woodward R, Wang PG, Feldman MF. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem. 2008:283:34596–34604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci U S A. 2005:102:3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geno KA, Gilbert Gwendolyn L, Song Joon Y, Skovsted Ian C, Klugman Keith P, Jones C, Konradsen Helle B, Nahm MH. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev. 2015:28:871–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner Carmen L, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. 2012:76:740–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GlaxoSmithKline . 2022. GSK 2022 infectious disease pipeline. https://www.gsk.com/en-gb/research-and-development/our-pipeline/?infectious-diseases.
- Harding CM, Feldman MF. Glycoengineering bioconjugate vaccines, therapeutics, and diagnostics in E. coli. Glycobiology. 2019:29:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CM, Nasr MA, Kinsella RL, Scott NE, Foster LJ, Weber BS, Fiester SE, Actis LA, Tracy EN, Munson RS Jr et al. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol. 2015:96:1023–1041. [DOI] [PubMed] [Google Scholar]
- Harding CM, Nasr MA, Scott NE, Goyette-Desjardins G, Nothaft H, Mayer AE, Chavez SM, Huynh JP, Kinsella RL, Szymanski CM et al. A platform for glycoengineering a polyvalent pneumococcal bioconjugate vaccine using E. coli as a host. Nat Commun. 2019:10:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney DJ, Hutchison AT, Hatchwell L, Humphrey SJ, James DE, Hocking S, Heilbronn LK, Larance M. Proteomic analysis of human plasma during intermittent fasting. J Proteome Res. 2019:18:2228–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H, Habash M, Aidoo F, Burrows LL. Single-residue changes in the C-Terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J Bacteriol. 2009:191:6513–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H, Bondy-Denomy J, Marquis H, Sztanko KM, Davidson AR, Burrows LL. Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat Microbiol. 2018:3:47–52. [DOI] [PubMed] [Google Scholar]
- Hayes AJ, Lewis JM, Davies MR, Scott NE. Burkholderia PglL enzymes are Serine preferring oligosaccharyltransferases which target conserved proteins across the Burkholderia genus. Commun Biol. 2021:4:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzempa J, Comer JE, Davis SA, Castric P. Glycosylation substrate specificity of Pseudomonas aeruginosa 1244 pilin. J Biol Chem. 2006:281:1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SJ, Karayel O, James DE, Mann M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat Protoc. 2018:13:1897–1916. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res. 2006:5:988–994. [DOI] [PubMed] [Google Scholar]
- Johnson & Johnson . 2022. Johnson & Johnson infectious diseases and vaccines, Global Public Health Pipeline; https://www.investor.jnj.com/pharmaceutical-pipeline-information. [Google Scholar]
- Kay EJ, Yates LE, Terra VS, Cuccui J, Wren BW. Recombinant expression of Streptococcus pneumoniae capsular polysaccharides in Escherichia coli. Open Biol. 2016:6:150243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Cuccui J, Wren BW. Recent advances in the production of recombinant glycoconjugate vaccines. NPJ Vaccines. 2019:4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoot CJ, Robinson LS, Harding CM. A minimal sequon sufficient for O-linked glycosylation by the versatile oligosaccharyltransferase PglS. Glycobiology. 2021:31:1192–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017:14:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop RM, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995:166:175–176. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014:11:319–324. [DOI] [PubMed] [Google Scholar]
- Lewis JM, Coulon PML, McDaniels TA, Scott NE. The application of open searching-based approaches for the identification of acinetobacter baumannii O-linked glycopeptides. J Vis Exp. 2021. [DOI] [PubMed] [Google Scholar]
- Lin C-w, Haeuptle MA, Aebi M. Supercharging reagent for enhanced liquid chromatographic separation and charging of sialylated and high-molecular-weight glycopeptides for nanoHPLC–ESI-MS/MS analysis. Anal Chem. 2016:88:8484–8494. [DOI] [PubMed] [Google Scholar]
- Liu B, Furevi A, Perepelov AV, Guo X, Cao H, Wang Q, Reeves PR, Knirel YA, Wang L, Widmalm G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev. 2020:44:655–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Forest K, Béretti J-L, Tainer J, Nassif X. Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Mol Microbiol. 1998:27:705–715. [DOI] [PubMed] [Google Scholar]
- Musumeci MA, Faridmoayer A, Watanabe Y, Feldman MF. Evaluating the role of conserved amino acids in bacterial O-oligosaccharyltransferases by in vivo, in vitro and limited proteolysis assays. Glycobiology. 2014:24:39–50. [DOI] [PubMed] [Google Scholar]
- Neal BL, Brown PK, Reeves PR. Use of Salmonella phage P22 for transduction in Escherichia coli. J Bacteriol. 1993:175:7115–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LC, Taguchi F, Tran QM, Naito K, Yamamoto M, Ohnishi-Kameyama M, Ono H, Yoshida M, Chiku K, Ishii T et al. Type IV pilin is glycosylated in Pseudomonas syringae pv. tabaci 6605 and is required for surface motility and virulence. Mol Plant Pathol. 2012:13:764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010:8:765–778. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Alpi E, Wang R, Hermjakob H, Vizcaino JA. Making proteomics data accessible and reusable: current state of proteomics databases and repositories. Proteomics. 2015:15:930–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019:47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto V, Berti F. Exploring the Group B Streptococcus capsular polysaccharides: the structural diversity provides the basis for development of NMR-based identity assays. J Pharm Biomed Anal. 2014:98:9–15. [DOI] [PubMed] [Google Scholar]
- Polasky DA, Yu F, Teo GC, Nesvizhskii AI. Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco. Nat Methods. 2020:17:1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstendörfer D, Gohl O, Mayer F, Averhoff B. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. strain BD413: regulation, modification, and cellular localization. J Bacteriol. 2000:182:3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutyan M, Henkel M, Horzempa J, Quinn M, Castric P. Glycosylation of pilin and nonpilin protein constructs by Pseudomonas aeruginosa 1244. J Bacteriol. 2010:192:5972–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002:71:635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007:2:1896–1906. [DOI] [PubMed] [Google Scholar]
- Rose CM, Rush MJ, Riley NM, Merrill AE, Kwiecien NW, Holden DD, Mullen C, Westphall MS, Coon JJ. A calibration routine for efficient ETD in large-scale proteomics. J Am Soc Mass Spectrom. 2015:26:1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan X, Loyola DE, Marolda CL, Perez-Donoso JM, Valvano MA. The WaaL O-antigen lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases. Glycobiology. 2012:22:288–299. [DOI] [PubMed] [Google Scholar]
- Saba J, Dutta S, Hemenway E, Viner R. Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. Int J Proteomics. 2012:2012:560391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer C, Messner P. Emerging facets of prokaryotic glycosylation. FEMS Microbiol Rev. 2017:41:49–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Kinsella RL, Edwards AV, Larsen MR, Dutta S, Saba J, Foster LJ, Feldman MF. Diversity within the O-linked protein glycosylation systems of Acinetobacter species. Mol Cell Proteomics. 2014:2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006:1:2856–2860. [DOI] [PubMed] [Google Scholar]
- Tan Rommel M, Kuang Z, Hao Y, Lee F, Lee T, Lee Ryan J, Lau Gee W, McCormick BA. Type IV pilus glycosylation mediates resistance of Pseudomonas aeruginosa to opsonic activities of the pulmonary surfactant protein A. Infect Immun. 2015:83:1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik Å, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci. 2009:106:4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Perry MB, MacLean LL, Yu SH. Structural analysis of the O-antigen side chain polysaccharides in the lipopolysaccharides of Klebsiella serotypes O2(2a), O2(2a,2b), and O2(2a,2c). J Bacteriol. 1992:174:4913–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks SJ, Denman C, Cia F, McCarthy E, Cuccui J, Wren BW. Virulence of the emerging pathogen, Burkholderia pseudomallei, depends upon the O-linked oligosaccharyltransferase, PglL. Future Microbiol. 2020:15:241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlieva L, Fülleborn JA, Walvoort MTC. Opportunities and challenges of bacterial glycosylation for the development of novel antibacterial strategies. Front Microbiol. 2021:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.




