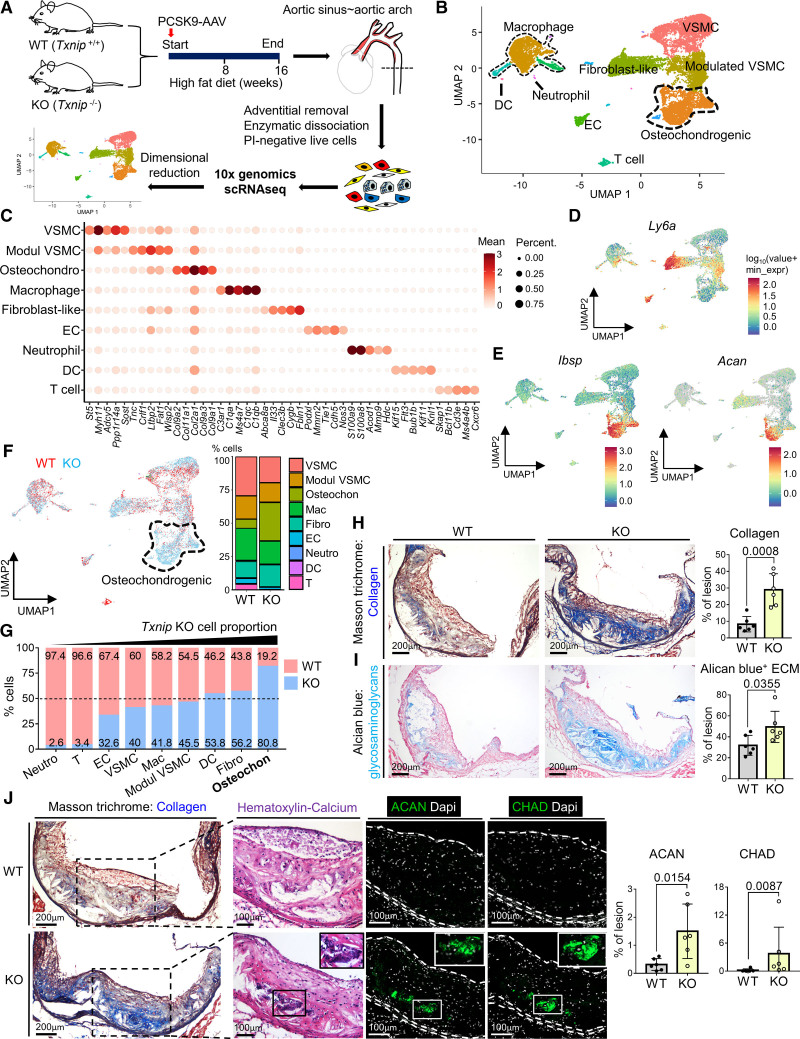
Figure 2.
Single-cell RNA-sequencing (scRNA-seq) analysis reveals the osteochondrogenic cluster enriched with bone- and cartilage-related genes, which is markedly expanded in Txnip knockout mice (Txnip KO) mice. A, Overall experimental procedure. The atherosclerotic plaques of 4 mice from each wild type (WT) and Txnip KO were pooled. After adventitial removal, the atherosclerotic lesion cells were enzymatically dissociated, and propidium iodide (PI)-negative cells were subjected to scRNA-seq. B, Uniform manifold approximation and projection (UMAP) visualization of integrated WT and Txnip KO scRNA-seq data. C, Top 5 differentially expressed genes (DEG) for each cluster. D and E, Feature plots for Ly6a, osteogenic marker Ibsp, and chondrogenic marker Acan (aggrecan). F, UMAP and cluster-annotated bar charts showing distributions of WT and Txnip KO cell. G, The bar graphs showing the proportions of WT and Txnip KO for each cluster. H and I, Representative serial sections and quantification results of Masson trichrome staining (H, for collagen), Alcian blue (I, for glycosaminoglycan extracellular matrix [ECM]) on the aortic sinus of WT and Txnip KO. n=6/6 for WT/Txnip KO. J, Representative serial section images showing the Masson trichrome staining, Hematoxylin and Eosin (H&E) staining, and immunostaining of the ACAN and CHAD (chondroadherin) on the aortic sinus sections of WT and Txnip KO. Masson trichrome staining, ACAN, and CHAD immunostainings are sequentially performed on 7 um of serial cryosections. H&E staining was performed on the CHAD-stained slides by removing the coverslips. ACAN and CHAD-positive areas were quantified (n=6/6 for WT/Txnip KO). The applied statistical tests and results are summarized in the Table S11. The error bars denote standard deviation. The exact P values are specified. Dapi indicates 4’‚6-diamidino-2-phenylindole; DC, dendritic cells; EC, endothelial cells; and Modul VSMC, modulated vascular smooth muscle cells.