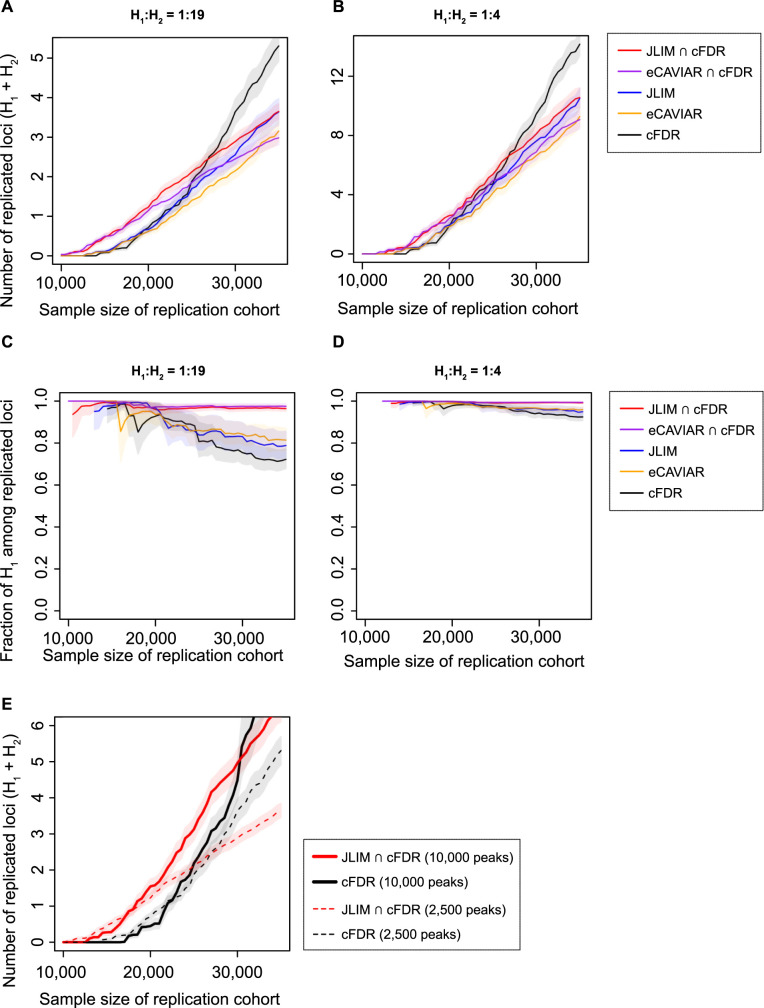
Fig 1. The projected number of replicated loci and proportion of shared causative variants among the replicated loci.
A total of 2,500 association peaks from well-powered GWAS studies (n = 150,000) were tested for the shared effect in simulated discovery cohorts (n = 10,000), and then select loci were tested for replication in simulated validation cohorts of the same genetic ancestry (n = 10,000–35,000). The candidate loci were identified by conditional false discovery rate (cFDR), colocalization tests (JLIM and eCAVIAR), or the intersection of cFDR and colocalization test. The p-value cutoff was set to 0.01 for cFDR and JLIM, and for eCAVIAR, the equivalent posterior threshold was calibrated using H0 simulation data. The 2,500 GWAS peaks consist of the loci simulating no causal effect for underpowered traits (H0) and those simulating the same causal effect between two traits (H1) or distinct causal effects (H2). The H1:H2 ratio was set to 1:19 (A and C) or 1:4 (B and D). The effect sizes of causative variants are correlated (ρ = 0.7) under H1 but uncorrelated under H2. Bonferroni correction was applied on replication tests. In Panel E, we contrast the number of replicated loci expected in Panel A (dashed line) with a more extreme scenario (10,000 GWAS peaks, consisting of 2,500 with H1:H2 of 1:19 and 7,500 with H1:H2 of 1:39; solid lines). In all, the proportion of H0 was set to 30% of all examined loci. The shaded area denotes the 95% CIs.