Figure 4.
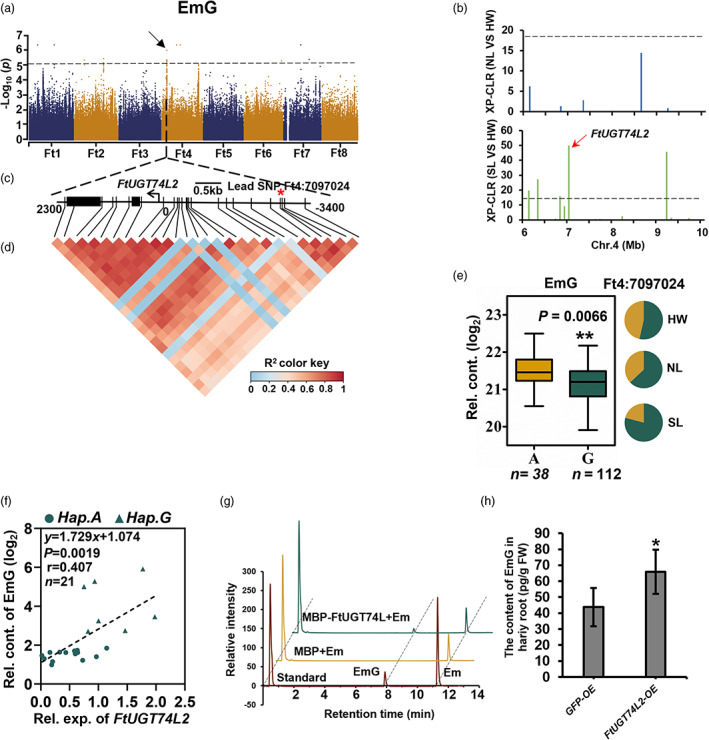
Decrease of emodin during domestication and identification of the biosynthetic enzyme FtUGT74L2. (a) Manhattan plot and QQ plot display the GWAS results of EmG content. The dashed line indicates the threshold −log10 P = 5. (b) The cross population composite likelihood ratio (XP‐CLR) between NL and HW (upper), and between SL and HW (lower) on chromosome 4. The black dashed horizontal lines indicate top 10% threshold for entire chromosome 4 (threshod with 18 in NL/HW and 14 in SL/HW). The red arrows indicate the position of FtUGT74L2 in the sweeps. (c) Gene model of FtUGT74L2. Filled black boxes represent coding sequence. The black vertical lines mark the polymorphic sites identified by high‐throughput sequencing, and the star (*) represents the lead SNP Ft4:7097024, located in the −3336 bp upstream FtUGT74L2. (d) A representation of pairwise r 2 value (a measure of LD) among all polymorphic sites in approximately 5.6 kb region around the lead SNP. The colour gradation of each box corresponds to the r 2 value according to the legend. (e) Frequencies of the high/low‐level trait‐related allele in three groups. Box plot indicates the relative content of EmG, plotted as a function of genotypes at SNP Ft4:7097024. The EmG content was log2 transformed, nHap.A = 38, nHap.G = 112. (f) Correlation between the relative contents of EmG and the transcription level of FtUGT74L2 in 21 buckwheat varieties, including 7 A‐hap and 14 G‐hap. P value was calculated using Student's t‐test. (g) FtUGT74L2 enzymatic assay in vitro. The different reaction curves by LC–MS were showed. Protein extract from E. coli containing the pMAL‐C5X empty vector was used as a negative control. (h) EmG content in FtUGT74L2 overexpression hairy roots mixed from three independent lines and three independent experiments were performed. The hairy roots of infected by A4 containing 35S: GFP(GFP‐OE) were used as negative control. Data in (e), (f) and (h) are presented as mean ± SD. Significant differences are indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student's t‐test.
