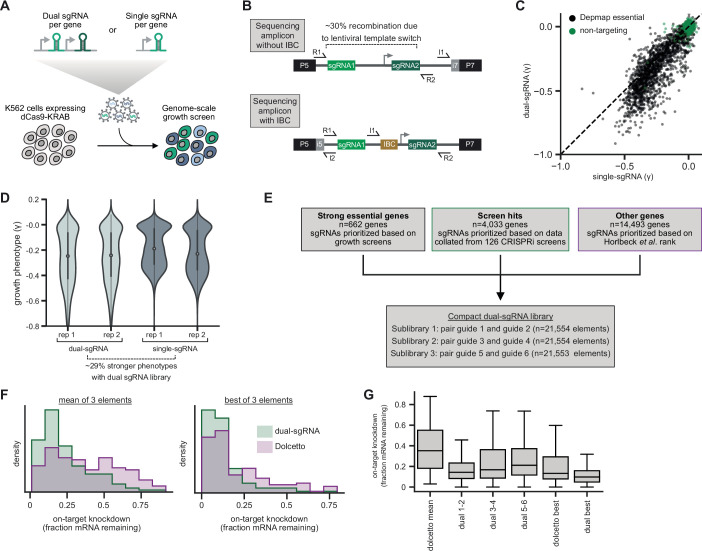
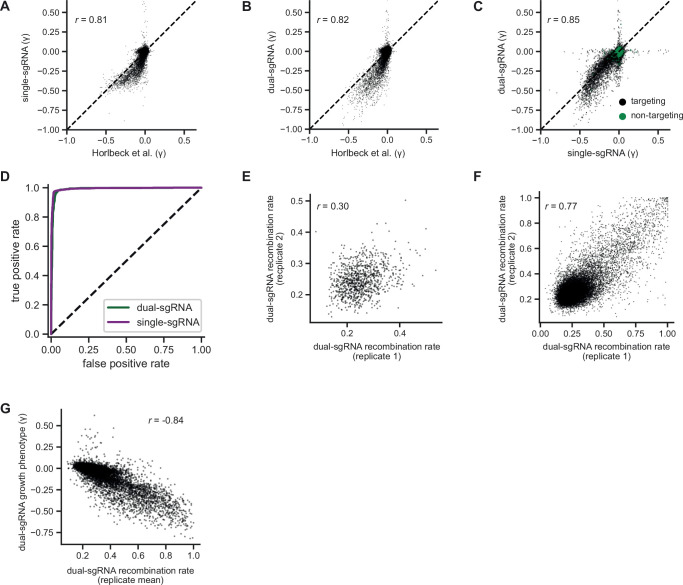
Figure 1. Design and validation of ultra-compact dual-single guide RNA (sgRNA) CRISPR interference (CRISPRi) libraries.
(A) Schematic of growth screen used to compare single- and dual-sgRNA libraries. (B) Schematic of dual-sgRNA library sequencing strategies. (C) Comparison of growth phenotypes for DepMap essential genes between single- and dual-sgRNA libraries. Sequencing libraries were prepared using the strategy labeled ‘Sequencing amplicon without IBC’ in panel B. Growth phenotypes are reported as γ (log2 fold-enrichment of Tfinal over T0, per doubling) and well correlated between libraries (r=0.91). Only values between –1 and 0.1 are shown. (D) Comparison of growth phenotypes for DepMap essential genes between single- and dual-sgRNA libraries. In the violin plot, the violin displays the kernel density estimate, the central white point represents the median, and the central black bar represents the interquartile range (IQR). (E) Design of final dual-sgRNA library. (F) Comparison of target gene knockdown by dual-sgRNA library versus Dolcetto library. Target gene knockdown was measured by single-cell RNA-sequencing (Perturb-seq). For each library, the ‘mean of 3 elements’ was calculated as the mean knockdown of all three elements targeting each gene. The ‘best of 3 elements’ represents the element with the best knockdown per each gene. (G) Comparison of target gene knockdown across elements in dual-sgRNA library versus Dolcetto. In the box plot, the box shows the IQR, the line dividing the box shows the median value, and the whiskers extend to show 1.5× the IQR. Outlier observations >1.5× IQR are not shown.