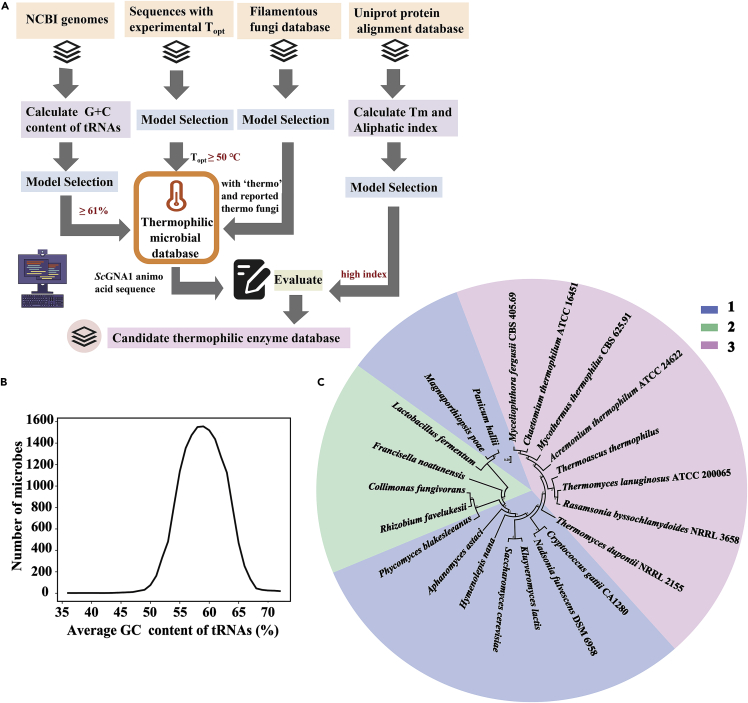
Figure 2.
Model development for mining thermophilic GNA1s with two strategies
(A) Schematic overview of the process to build candidate thermophilic enzyme database. Topt, the optimal growth temperatures of organisms.
(B) Variation curve of microbial numbers with NCBI genomic average GC content of tRNAs (%).
(C) Phylogenetic tree analysis of the 20 selected candidate GNA1 thermophilic enzymes. 1. Based on Uniprot alignment according to ScGNA1 amino acid sequence. 2. Based on the GC content of tRNAs in microbial genomes and reported optimal growth temperatures of organisms. 3. Based on the thermophilic fungus database.