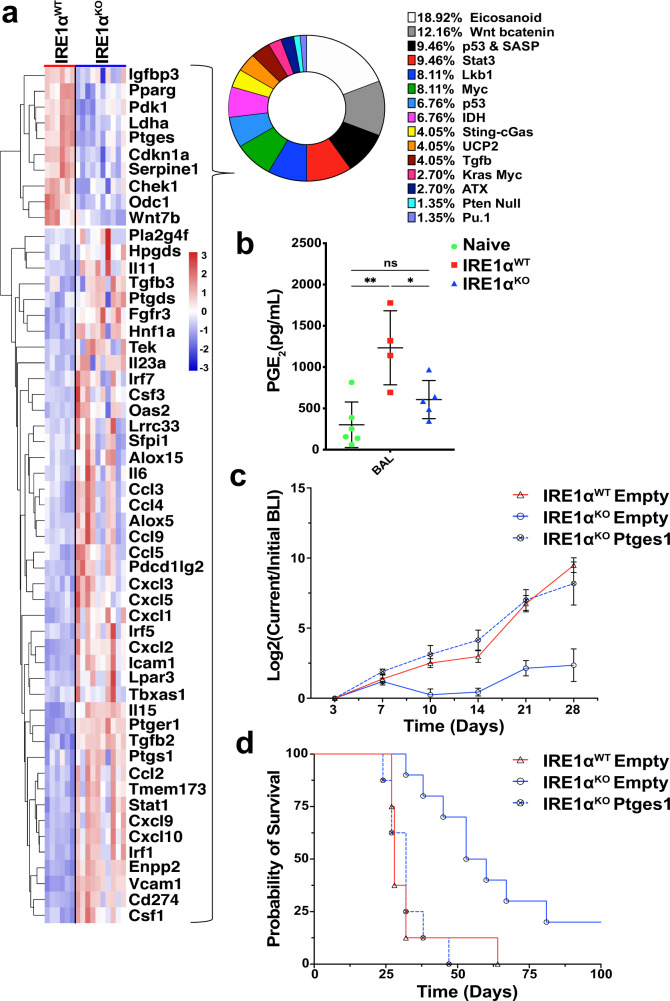
Fig. 4. IRE1α-XBP1 signaling drives immunosuppressive PGE2 production that promotes NSCLC progression.
a Heatmap of differentially expressed genes between IRE1αKO vs. IRE1αWT HKP1 cells from the Immunomodulator database. b BAL PGE2 levels as measured by ELISA from tumor naïve (green, n = 6), and IRE1αWT (red, n = 4), or IRE1αKO (blue, n = 5) tumor-bearing lungs. Data were shown as mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test for ratios; *P < 0.05, **P < 0.001, and ***P < 0.0001. c BLI plots of longitudinally tracked in vivo IRE1αWT empty vector (red, n = 10), IRE1αKO empty vector (blue, n = 10) and IRE1αKO Ptges cDNA (blue broken line, n = 10) HKP1 tumors. Data were shown as mean ± SEM of biological replicates. Analyses of different time points in tumor progression were performed using two-way ANOVA with Tukey’s multiple comparisons test; *P < 0.05, **P < 0.001, and ***P < 0.0001. d Kaplan–Meier plots showing the probability of overall survival in IRE1αWT (red) vs IRE1αKO (blue) and IRE1αKO Ptges cDNA HKP1 tumor-bearing mice (P < 0.001). Tumors were allowed to progress until the endpoint and survival were evaluated using Mantel–Haenszel Log-rank-test). Source data are provided as a Source Data file.