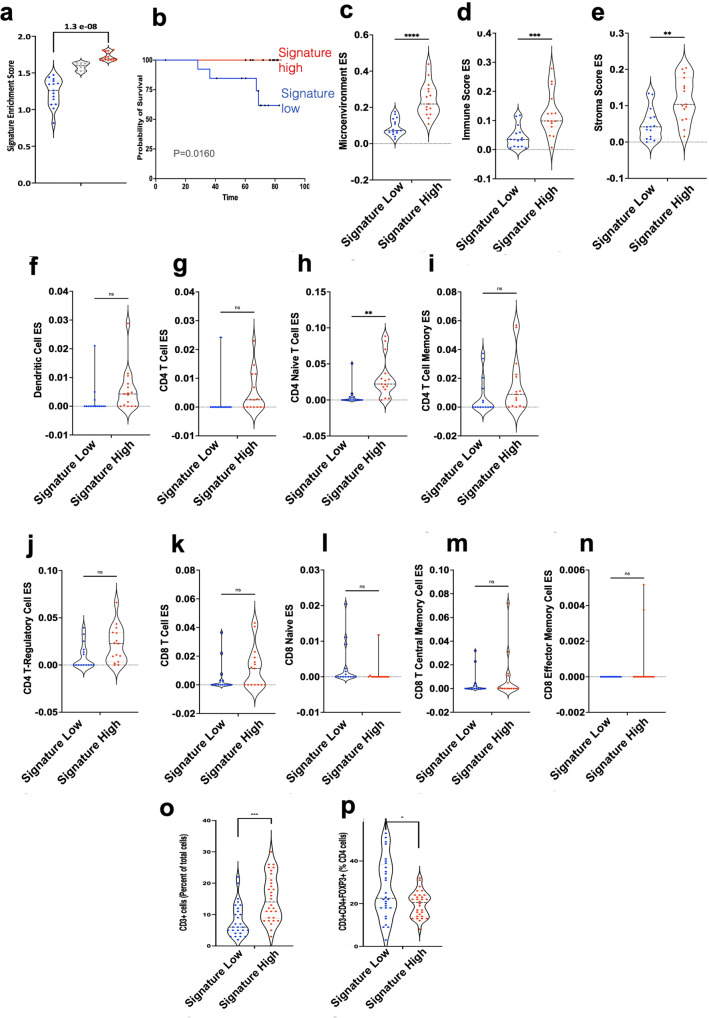
Fig. 6. Validation of the IRE1a signature and RNA-seq deconvolution.
a Violin plot of enrichment scores (ssGSEA) for IRE1αKO gene signature (Log2 fold-change >1 and FDR 0.01) low (n = 15, blue), mid (gray, n = 14), and high (red, n = 15) NSCLC patients. One-way ANOVA with Tukey’s multiple comparisons test for ratios. b Kaplan–Meier survival plots depicting associations between overall survival (OS) and IRE1αKO signature status of NSCLC patients stratified into the top and bottom tertiles (15 patients in each) for the IRE1aKO gene signature. c–n Violin plots of xCell pipeline enrichment scores for microenvironment (c P < 0.0001), immune (d P = 0.001), stromal (e P = 0.001), dendritic (f P = ns), CD4 + T cells (g P = ns), CD4 + naïve (h P = 0.0027), CD4 memory (i P = ns), T-regulatory (j P = NS), CD8 + T cell (k P = ns), CD8 + Naïve (l P = ns), CD8 + TCM (m P = ns), and CD8 + effector memory (n P = ns) from the validation set of lung cancer patients from the IRE1α signature high (red, n = 15) and low (blue, n = 15) groups for the enrichment of various immune cells. Unpaired, two-tailed, Student’s t-test. *P < 0.05, **P < 0.001, and ***P < 0.0001. o, p IF analysis showing quantitation of CD3T cells (left panel) and Tregs (right panel) in tumor nests of validation set of lung cancer patients from the IRE1α signature high (red, n = 30) and low (blue, n = 30) groups. Unpaired, two-tailed, Student’s t-test. *P < 0.05, **P < 0.001, and ***P < 0.0001. Source data are provided as a Source Data file.