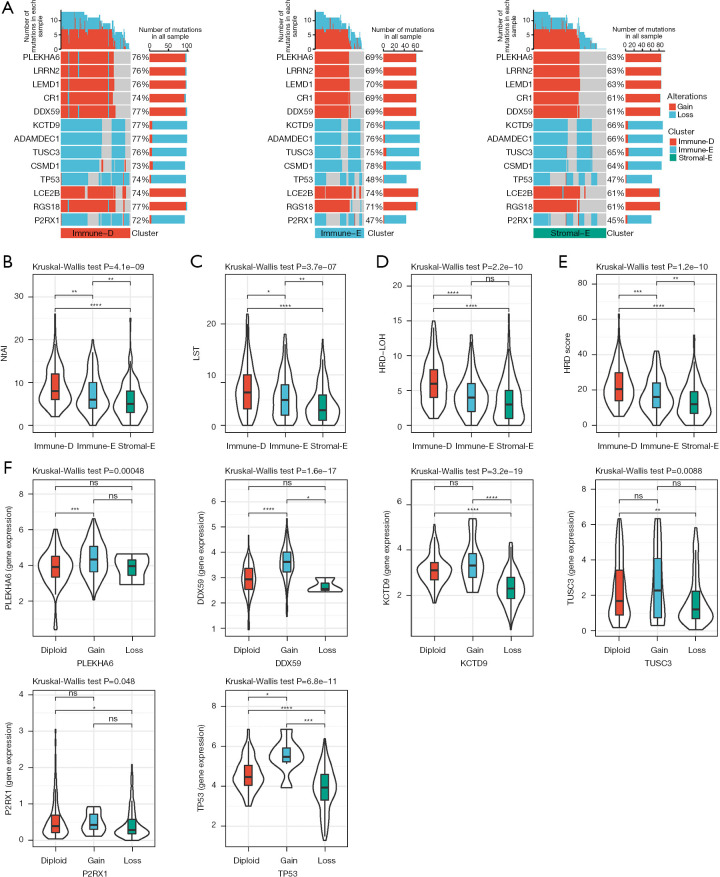
Figure 5.
Comparison of genomic alterations in three clusters in TCGA dataset. (A) The CNVs of partial genes in 3 clusters. (B-E) The scores of NtAI, LST, HRD-LOH, and HRD in 3 clusters. Kruskal-Wallis test was conducted. (F) The expression of diploid, CNV gain and CNV loss groups in 6 genes. Kruskal-Wallis test was conducted. ns, not significant; *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. NtAI, number of telomeric regions with allelic imbalance; LST, large-scale transition; HRD, homologous recombination defect; LOH, loss of heterozygosity; TCGA, The Cancer Genome Atlas; CNV, copy number variation.