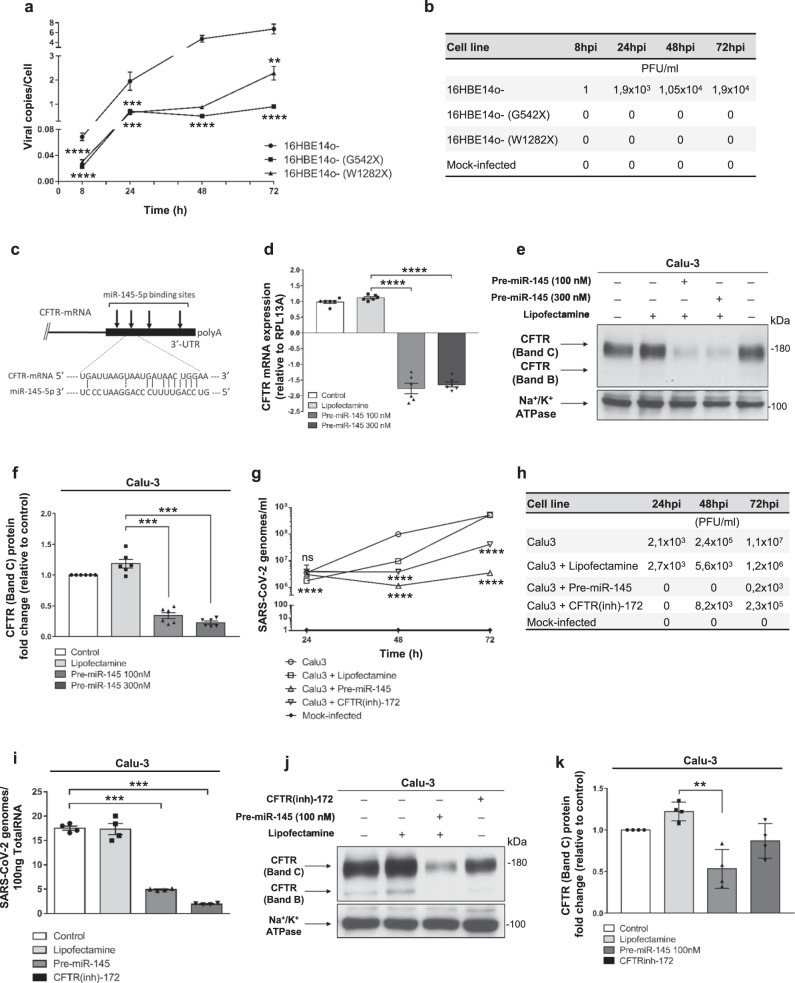
Fig. 6. Loss of CFTR expression and function inhibits SARS-CoV-2 entry and replication.
a 16HBE14o- and mutant clones carrying W1282X- and G542X-CFTR were infected with 0.1 MOI SARS-CoV-2 for 1 h, and then the viral copies/cell in cell lysates was evaluated 8, 24, 48, and 72 hpi by dPCR. The results are shown as the mean ± SEM from three independent experiments (n = 3). b Viral titration was performed on supernatants of 16HBE14o- cells and their mutant clones infected with 0.1 MOI SARS-CoV-2 or 0.1 moi UV-inactivated SARS-CoV-2 (mock) for 1 h, and then the viral titration was performed by plaque assay after 8, 24, 48, and 72 hpi. c Location of the miR-145-5p binding sites within the CFTR 3′-UTR (binding site displaying the highest affinity is shown). d–f Effect of premiR-145-5p treatment on CFTR expression in Calu-3 cells. The effect was verified by RT–qPCR (d) and western blot analysis (e) in six independent experiments (n = 6). f Densitometry analysis of six independent experiments (n = 6), conducted as reported in (e). Data represented in panels d–f are shown as the mean ± SEM. Effect of premiR-145-5p compared to CFTR(inh)−172 on the extracellular release of SARS-CoV-2 in infected Calu-3 cells. Extracellular release of SARS-CoV-2 genomes (g); viral titration has been performed in Calu-3 cells upon SARS-CoV-2 infection sustained for 24, 48, and 72 hpi (h); intracellular production of SARS-CoV-2 genomes (i). Results are reported as the mean ± SEM from four independent experiments (n = 4). Western blot analysis of CFTR (j) and corresponding densitometry analysis (k) in response to SARS-CoV-2 infection under the same conditions depicted in panels (g–i). Results are reported as the mean ± SEM from four independent experiments (n = 4). Normal distribution was tested by the Shapiro–Wilk test before running the two-tailed Student’s t test for paired data (a, d, f, g, i, k), which has been reported within the scatter plot with bars (**p < 0.01; ***p < 0.001; ****p < 0.0001). In panels a and g, for some points, the error bars were shorter than the height of the symbols. In this case, the error bars were not drawn. Source data are provided as a Source Data File.