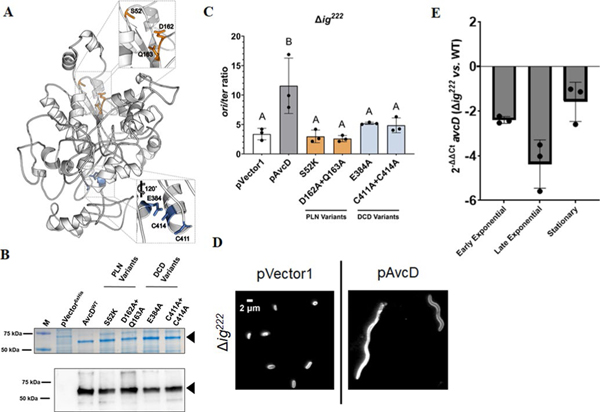
Extended Data Fig. 8 |. Mutations in conserved residues of AvcD do not affect the stability or function of the protein.
(A) Phyre2 (ref. 18) predicted structure of AvcD from V. cholerae El Tor. Insets highlight conserved residues of the PLN (top) and DCD (bottom) domains selected for mutagenesis. (B) Representative Coomassie stained gel (top) and anti-6x His antibody Western blot (bottom) of whole cell lysates from E. coli BL21(DE3) cells maintaining an empty vector (pVector6xHis), inducible C-terminal 6x histidine tagged avcD (WT) or avcD variants (S52K, D162A + Q163A, E384A, and C411A + C414A) grown in the presence of 1 mM IPTG for 3 h. Sample inputs were normalized by culture OD600 and resolved by SDS-PAGE. Three biological replicates of each strain were analyzed with similar results. Black triangles correspond to the predicted molecular weight of the AvcD tagged fusions (60.6 kDa). M = molecular weight marker. (C) V. cholerae mutant expressing the indicated AvcD variants. ori/ter ratios of Chromosome 1 in Δig222 V. cholerae strains expressing the indicated pAvcD construct and quantified using qRT-PCR. Each bar represents the mean ± SEM, n=3. Different letters indicate significant differences (n=3) at p < 0.05, according to Two-way ANOVA with Tukey’s post-hoc test. (D) Representative images of Δig222 cultures maintaining an empty vector plasmid pVector 1 or pAvcD grown in the presence of 100 μM IPTG for 8 h. Cells were stained with FM4–64 prior to imaging and performed in biological triplicate. (E) Relative difference in avcD expression between Δig222 and WT V. cholerae at three different growth phases using qRT-PCR and an endogenous gyrA control. Data represent the mean ± SEM of three biological replicates.