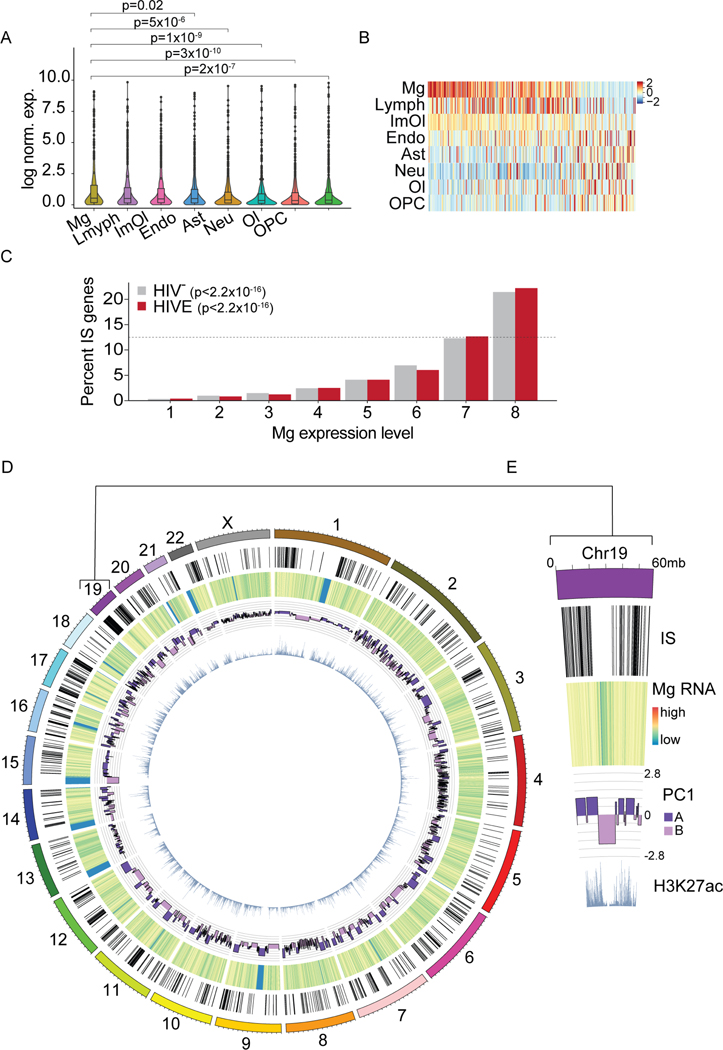
Figure 5. Viral integration is associated with microglial transcription and genome structure.
(A) snRNA-seq FC/WM violin plots showing cluster-specific expression of all genes containing IS in HIVE samples (n=7). For cell types composed of more than 1 subcluster, values were averaged across all cells. P, Wilcoxon rank sum (Mg vs. other cell types). (B) Heatmap showing expression centered and scaled by gene for top 300 IS genes most variably expressed across cell types. (C) Graph comparing IS gene expression levels in HIVE and HIV- microglia, divided into 8 equal groups (12.5% of genes/group, ranked by successively higher expression). Dotted line 12.5%. P, Poisson linear regression. (D-E) Genome-wide and chromosome 19 specific circos plot displaying, from the outside in, NeuN- IS, heatmap of log scaled HIVE Mg snRNA-seq read counts, HIVE Mg Hi-C PC1 value, and NeuN- H3K27ac ChIP-seq reads. See also Figure S6.