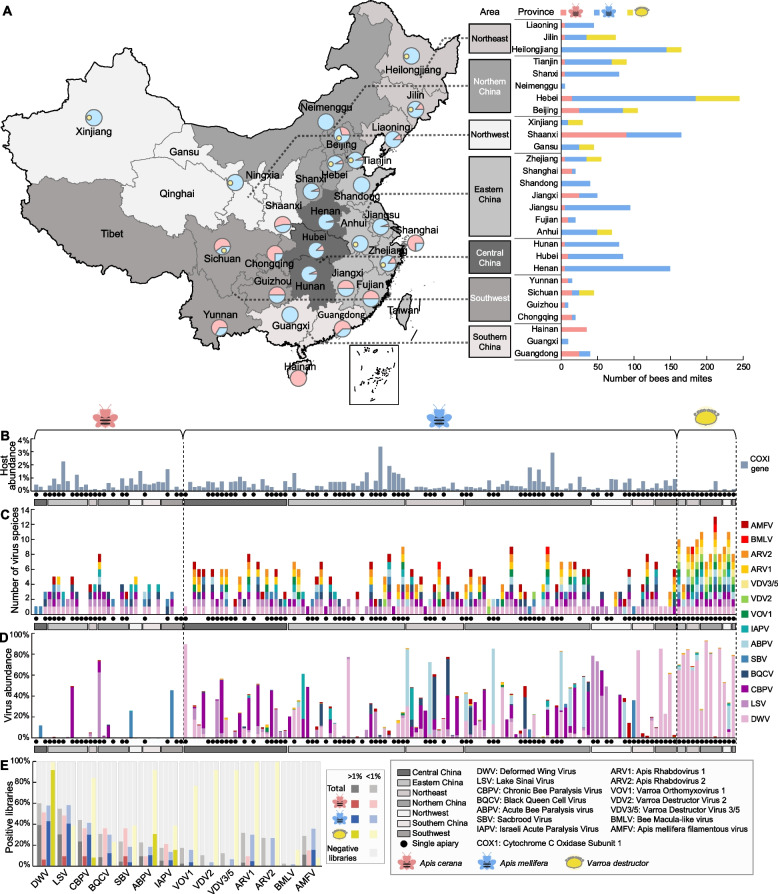
Fig. 1.
Identification of known honeybee viruses in China. A Sampling locations in China (left) and number of specimens surveyed in each host species in individual provinces (right), colored by host types. B Percentage of non-rRNA reads aligning to the honeybee and mite mitochondrial COX1 reference sequences. C Number of known viral species identified in each library, colored by virus species. D Percentage of non-rRNA reads mapped to each RNA virus genomes in each library, colored by virus species. E Percentage of positive libraries for each virus, which was further subdivided at either >1% non-rRNA reads (dark color) or <1% non-rRNA reads (light color) within different host species