Abstract
Although the risk of SARS-CoV-2 transmission through lung transplantation from acutely infected donors is high, the risks of virus transmission and long-term lung allograft outcomes are not as well described when using pulmonary organs from COVID-19–recovered donors. We describe successful lung transplantation for a COVID-19–related lung injury using lungs from a COVID-19–recovered donor who was retrospectively found to have detectable genomic SARS-CoV-2 RNA in the lung tissue by multiple highly sensitive assays. However, SARS-CoV-2 subgenomic RNA (sgRNA), a marker of viral replication, was not detectable in the donor respiratory tissues. One year after lung transplantation, the recipient has a good functional status, walking 1 mile several times per week without the need for supplemental oxygen and without any evidence of donor-derived SARS-CoV-2 transmission. Our findings highlight the limitations of current clinical laboratory diagnostic assays in detecting the persistence of SARS-CoV-2 RNA in the lung tissue. The persistence of SARS-CoV-2 RNA in the donor tissue did not appear to represent active viral replication via sgRNA testing and, most importantly, did not negatively impact the allograft outcome in the first year after lung transplantation. sgRNA is easily performed and may be a useful assay for assessing viral infectivity in organs from donors with a recent infection.
Keywords: SARS-CoV-2, COVID-19, lung transplant, subgenomic RNA, donor-derived infection
1. Introduction
As the COVID-19 pandemic continues, an increasing number of potential organ donors will have active COVID-19 or recent illness or would have recovered from a more remote COVID-19 infection before organ donation. Early in the pandemic, recovery of organs from donors with COVID-19 was discouraged because of the systemic nature of COVID-19 manifestations. SARS-CoV-2 viral RNAemia was demonstrated in up to 15% of COVID-19 cases, and the viral genomic material was detected in extrapulmonary tissues.1, 2, 3 This raised concerns about the potential for donor-derived viral transmission through organ transplantation. Data now suggest that transplanting nonpulmonary organs from donors with a resolved SARS-CoV-2 infection is unlikely to result in viral transmission to recipients.4 , 5 More importantly, transplanting nonpulmonary organs from donors who are actively positive for SARS-CoV-2 at the time of organ procurement also appears safe, with no reported viral transmission to date.6 , 7 Accordingly, the number of organs recovered from SARS-CoV-2–positive donors has been increasing, with 178 nonpulmonary organs transplanted from 66 SARS-CoV-2–positive donors between May 27, 2021, and November 30, 2021.8
It is presumed that the risk of SARS-CoV-2 transmission through lung transplantation is higher, primarily because SARS-CoV-2 tropism is highest in the respiratory tract, mediated by the high density of angiotensin converting enzyme-2 receptors.9 There have been 2 published cases of SARS-CoV-2 transmission through lung transplantation resulting in respiratory failure.10 , 11 In both cases, the donors were not known to have COVID-19 and tested negative for SARS-CoV-2 by polymerase chain reaction (PCR) from a nasopharyngeal sample. At that time, SARS-CoV-2 testing of a lower respiratory tract (LRT) sample was not required before lung procurement, and in both cases, testing of archived donor LRT samples was positive for SARS-CoV-2. Subsequently, the Organ Procurement and Transplantation Network (OPTN) mandated SARS-CoV-2 nucleic acid amplification testing from donor LRT samples in all cases where lungs are procured.12
Although lung allografts from SARS-CoV-2–positive donors with an active infection should be avoided, the safety of pulmonary organs from donors who have recovered from COVID-19 is not well described. Because SARS-CoV-2 RNA can be shed in the respiratory tract for weeks despite symptomatic improvement,13, 14, 15 it is difficult to determine the optimal time to recover lungs from donors with a recent COVID-19 infection. This is a critical question because the exclusion of these donors could restrict an already limited resource, and the COVID-19–recovered status is increasingly prevalent. To date, only 3 cases of lung transplantation involving COVID-19–recovered donors have been reported.16, 17, 18 Herein, we describe a successful lung transplantation for a COVID-19–related lung injury where the donor lung tissue was retrospectively found to have detectable SARS-CoV-2 RNA by multiple assays, including highly sensitive droplet digital PCR (ddPCR), viral sequencing, and in situ hybridization (ISH). However, subgenomic RNA (sgRNA) was not present despite the detection of genomic viral RNA in the donor tissue, suggesting the absence of replicating virus. There was no evidence of SARS-CoV-2 transmission to the recipient, and the recipient is doing well 1 year after transplantation. This case adds to the growing body of evidence demonstrating the potential safety of lung transplantation using COVID-19–recovered donors. We demonstrate some of the limitations of COVID-19 diagnostic assays used in the current clinical practice to detect viral persistence in lung tissues and highlight the potential role of sgRNA testing to exclude the presence of replication-competent virus when genomic RNA is detectable.
2. Case presentation
2.1. Recipient information
The lung transplant recipient was a 62-year-old man with a history of mild chronic obstructive pulmonary disease, chronic left hemidiaphragm paralysis, and pulmonary sarcoidosis. He was admitted to a community hospital with hypoxemic respiratory failure 7 days after testing positive for SARS-CoV-2. Before his COVID-19 diagnosis, he did not require supplemental oxygen. He was treated with remdesivir and dexamethasone with gradual improvement and was discharged home 13 days after his initial diagnosis, requiring 2 L of supplemental oxygen.
Thirty-one days after his initial diagnosis, the patient was readmitted with progressive respiratory failure. His oxygen saturation upon admission was 83% while breathing 2 L of O2 via a nasal cannula. He was placed on a 50-L high-flow nasal cannula with a fraction of inspired oxygen of 70%. He was treated for suspected bacterial pneumonia, and solumedrol was administered. Because of progressive hypoxic respiratory failure, he was transferred to our medical center, 40 days from his initial COVID-19 diagnosis, for an urgent lung transplant evaluation.
At the time of hospital transfer, the patient had a negative SARS-CoV-2 PCR test from a nasopharyngeal sample and had reactive antibodies to the SARS-CoV-2 spike protein. He completed an expedited transplant evaluation and was listed for lung transplantation 42 days after his initial COVID-19 diagnosis. He remained stable on the high-flow nasal cannula, and on day 56 after his initial COVID-19 diagnosis, suitable lungs became available.
2.2. Donor information
The donor was in the fourth decade of life and developed an anoxic brain injury with herniation in the setting of cardiac arrest from drug overdose, resulting in the declaration of brain death. The airway examination via bronchoscopy revealed normal anatomy and few-to-moderate secretions that cleared easily with suctioning. Chest computed tomography imaging demonstrated dependent consolidations consistent with possible aspiration pneumonia and bilateral pneumothoraces. The donor tested negative for SARS-CoV-2 by nucleic acid amplification testing on upper respiratory tract and LRT samples. The lungs were accepted for transplantation.
2.3. Lung transplantation and post-lung transplant course
The recipient underwent sequential bilateral lung transplantation using clamshell thoracotomy and was supported with central veno-arterial extracorporeal membrane oxygenation. He was decannulated from veno-arterial extracorporeal membrane oxygenation upon completion of the procedure. He received induction with methylprednisolone 1000 mg, and his maintenance immunosuppression therapy consisted of tacrolimus, mycophenolate mofetil, and prednisone. Routine bronchoscopy performed on posttransplant days 1 and 4 revealed intact bronchial anastomoses and mucoid secretions in the distal airways. He underwent tracheostomy on posttransplant day 6 because of the ongoing need for mechanical ventilatory support.
Over the next 2 weeks, he was gradually weaned from the mechanical ventilator support. He was discharged from the intensive care unit on posttransplant day 20 and his tracheostomy was decannulated on posttransplant day 26. The result of a lung biopsy performed on posttransplant day 25 was negative for acute cellular rejection. He was discharged home breathing ambient air on posttransplant day 32.
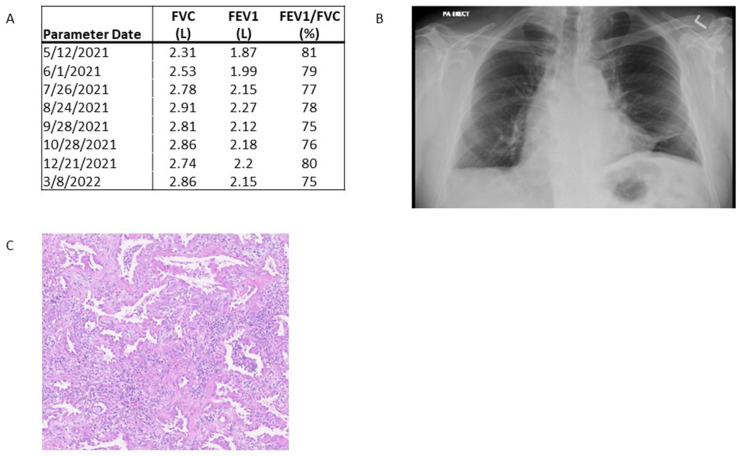
SARS-CoV-2 PCR test performed on a bronchoalveolar lavage (BAL) fluid sample from our recipient was negative on posttransplant day 1 and remained negative through the first 3 months after transplantation. The patient is now >1 year posttransplantation, is walking 1 mile approximately 4 times weekly without needing supplemental oxygen, and has a normal lung function (Fig. 1 A) and clear chest radiography (Fig. 1B).
Fig. 1.
Clinical data from lung transplant recipient. (A) Serial pulmonary function testing demonstrates improvement in forced expiratory volume (FEV1) and forced vital capacity (FVC) between 3 and 12 months post-lung transplantation. (B) Chest radiograph at 12 months after lung transplantation demonstrating clear lung parenchyma. (C) Hematoxylin and eosin staining of the recipient’s explanted lung tissue showing a diffuse alveolar injury with patchy organizing pneumonia.
3. Posttransplant analyses of donor and recipient tissues and blood
3.1. Lung histopathology
The evaluation of explanted lungs from our recipient demonstrated smoking-related changes with an apical bulla. There were microscopic findings of a diffuse alveolar injury and patchy organizing pneumonia (Fig. 1C).
3.2. Donor and recipient SARS-CoV-2 serology results
We evaluated pretransplant residual blood samples from both the donor and recipient for the presence of SARS-CoV-2–specific antibodies (Supplementary Methods). Immunoglobulin G (IgG) antibodies specific for SARS-CoV-2 nucleocapsid and spike proteins were detected in the plasma of the recipient, consistent with the recipient’s COVID-19 diagnosis before transplantation. We also detected IgG antibodies specific for both SARS-CoV-2 nucleocapsid and spike proteins in donor plasma—evidence that the donor had a prior history of COVID-19. The detection of IgG antibodies directed against SARS-CoV-2 spike proteins in both the donor and recipient was due to a natural infection as both these infections occurred before the widespread availability of SARS-CoV-2 vaccines.
3.3. SARS-CoV-2 RNA detected in recipient explants and donor tissues by ddPCR
To determine whether SARS-CoV-2 RNA was present in any fluid or tissue samples from the donor or recipient at the time of transplantation, ddPCR was performed on residual clinical samples (Supplementary Methods). SARS-CoV-2 RNA was not detectable in the plasma of the donor or the recipient at the time of transplantation. We did not detect SARS-CoV-2 RNA in the BAL fluid collected from donor lungs just before transplantation or in the BAL fluid collected from the recipient on posttransplant day 1 (Table 1 ).
Table 1.
Quantification of SARS-CoV-2 RNA in body fluids and tissues by ddPCR.
| Blood | BAL pre-/peritransplant | BAL posttransplant day 1 | Left lung |
Right lung |
Distal trachea | Left bronchus | Right bronchus intermedius | Lymph node |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upper lobe | Lower lobe | Upper lobe | Middle lobe | Lower lobe | Tracheal | Thoracic node 1 | Thoracic node 2 | |||||||
| Recipient/explant | – | – | – | 0.0104 | 0.044 | 0.0042 | 0.0108 | – | n/a | – | 0.0079 | n/a | n/a | n/a |
| Donor | – | – | n/a | n/a | n/a | 0.0099 | – | n/a | – | n/a | n/a | 0.0054 | – | 0.006 |
Detectable RNA is presented as the average nucleocapsid gene copies per ng of RNA added to the assay. “–” indicates a negative result under the limit of detection of the assay. “n/a” indicates that the tissue was not available for ddPCR.
BAL, bronchoalveolar lavage; ddPCR, droplet digital polymerase chain reaction.
We detected SARS-CoV-2 RNA using ddPCR in multiple thoracic tissues collected from both the donor and recipient at the time of transplantation (Table 1). We detected SARS-CoV-2 RNA in the left upper and lower lung lobes, right upper and middle lung lobes, and right bronchus intermedius collected from the recipient’s explanted tissue. From the donor, we detected SARS-CoV-2 RNA in the right upper lung lobe tissue and from 2 lymph nodes, which were removed during the preparation of the lung before implantation and slated for discard.
3.4. SARS-CoV-2 RNA detected in recipient explants and donor tissues by ISH
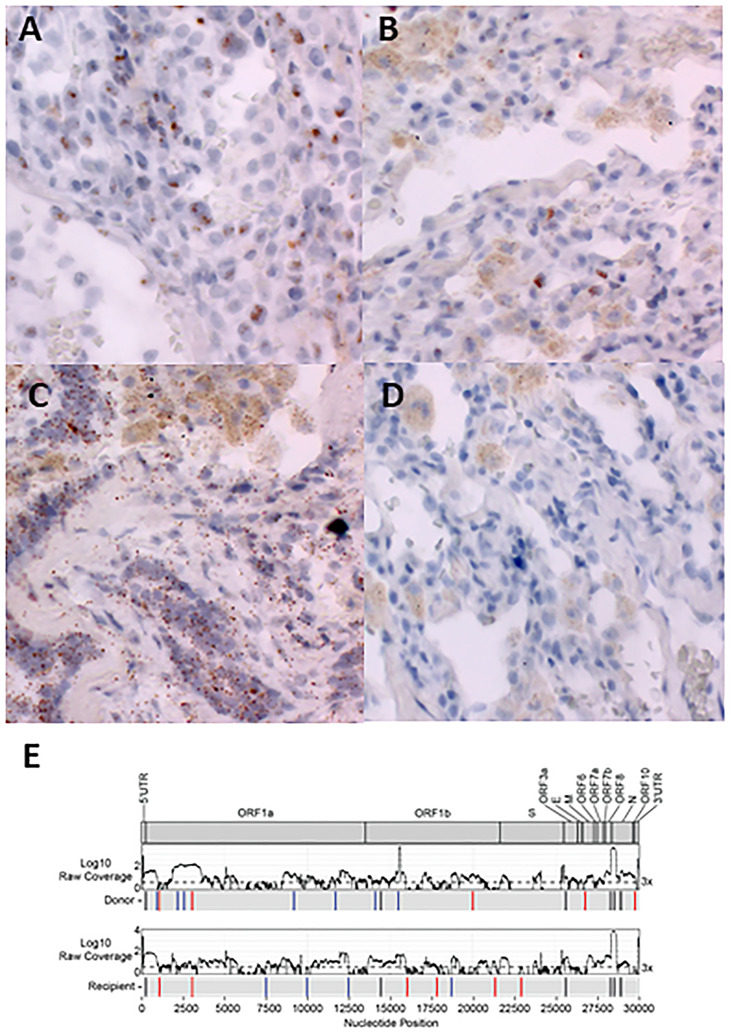
To substantiate the results of ddPCR and verify that viral RNA was in the tissue, RNA ISH (RNAscope, Advanced Cell Diagnostics) was performed to assess the presence of SARS-CoV-2 spike RNA on lung sections and to localize SARS-CoV-2 RNA to the cell level (Supplementary Methods). In the lung tissue explanted from the recipient, SARS-CoV-2 RNA was detected within the squamous metaplastic epithelium associated with diffuse alveolar damage (Fig. 2 A). Additionally, we detected the presence of SARS-CoV-2 RNA in the donor lung tissue obtained at the time of transplant within the alveolar epithelial cells (Fig. 2B). As a positive control for this assay, we also detected signal from a probe that targets RNA of a mammalian housekeeping gene (Fig. 2C). There was no signal detected from the negative control probe that targets bacterial RNA (Fig. 2D). Both the control probes were tested on the donor lung sample shown in Fig. 2B.
Fig. 2.
RNA in situ hybridization (RNAscope) and viral sequencing for the detection of SARS-CoV-2 in recipient and donor lung tissues. (A) Positive chromogenic signals (brown) from the probe for SARS-CoV-2 spike RNA on explanted recipient lung tissue. (B) Positive chromogenic signals (brown) from the probe for SARS-CoV-2 spike RNA on donor lung tissue. (C) Positive chromogenic signals (brown) from probe Hs-PPIB, a pan-mammalian housekeeping gene, on donor lung tissue (positive control for the assay). (D) No chromogenic signal from the probe for bacterial gene RNA on donor lung tissue (negative control for the assay). Original magnification for all images: ×40. (E) Donor and recipient lung samples display significant sequence diversity. Coverage plots (top) and the schematic (bottom) showing consensus changes across the SARS-CoV-2 genome in donor lung and recipient lung samples. Coverage is plotted as log10 of the raw read depth; dotted line is of 3× coverage. Bars in the schematic indicate regions where consensus changes are found in each sample, compared with the Wuhan/Hu-1 strain. Black bars show consensus changes present in both samples, blue bars show consensus changes unique to a single sample, red bars indicate unique consensus changes in which the coverage in the other sample does not reach the cutoff of 3×, and red open boxes indicate an identified consensus change that is present in <3 reads.
3.5. No SARS-CoV-2 sgRNA detected in donor or recipient lungs
To determine whether the presence of genomic viral RNA reflected an active viral infection, we assayed for sgRNA, a proposed surrogate marker of active virus replication (Supplementary Methods). sgRNA was not detected in the respiratory tissue from either the recipient or the donor.
3.6. Whole genome sequencing identified SARS-CoV-2 in both donor and recipient lung tissue
Whole genome sequencing was performed on donor and recipient lung tissue samples to further validate the presence of SARS-CoV-2 and to investigate sequence diversity (Supplementary Methods). SARS-CoV-2 sequences were identified in both donor and explanted recipient lung tissues. Because of the low viral load of the inputs, coverage across the genome was inconsistent, and lineage calling of the virus found in each sample was not possible. However, we identified several consensus changes that were unique to each sample, as well as some notable changes that were shared between the virus sequence found in the donor and recipient (Fig. 2E). Both viruses contained the ORF1b:P314L mutation, a defining mutation that separated the nextstrain 20A (pangolin lineage B.1) clade from 19A (pangolin lineage B), and the 28881:28883 GGG > AAC mutation (N:R203K, G204R), which is a defining mutation of the nextstrain 20B clade (pangolin lineage B.1.1). Each sample contained several unique changes as well, 10 in the donor sample and 8 in the recipient sample.
4. Discussion
Whether SARS-CoV-2 can be transmitted through lung transplantation from donors who have recovered from or had recent infection is uncertain and has resulted in reduced utilization of lungs from donors with recent COVID-19 infection. This appears driven partly by concerns about the persistence of SARS-CoV-2 viral RNA in the lung and extrapulmonary tissue, which can be detected for weeks to months after the initial diagnosis.13 , 15 , 19 However, the detection of genomic RNA cannot differentiate replicating viruses from nonreplicating viruses, resulting in discard of organs that may not pose a risk of viral transmission. An assay capable of detecting active viral replication with high sensitivity could prove useful in further stratifying the risk of SARS-CoV-2 transmission from donors with a recent COVID-19 infection.
In our case, SARS-CoV-2 was not transmitted to our lung transplant recipient despite the demonstrated persistence of genomic SARS-CoV-2 RNA in the donor lung tissue. SARS-CoV-2 RNA was detected by ddPCR, and its presence was confirmed using an RNAscope, which showed its localization to alveolar epithelial cells. To further ensure that SARS-CoV-2 RNA detected in the donor lung tissue was not caused by contamination from the recipient at the time of transplantation, we undertook virus sequencing analysis. Although limited coverage prevents the exact lineage identification, the sequencing confirmed the presence of SARS-CoV-2 RNA, and the diversity observed between the samples suggests that the viruses in the 2 individuals were possibly from different lineages.
Although RNAscope can detect the presence of viral transcripts in tissue samples and was utilized by several groups to confirm the absence of SARS-CoV-2 before organ transplantation,16 , 20 sensitive assays for genomic RNA, such as RNAscope and ddPCR, cannot determine whether these transcripts are from the replicating virus or are remnant transcripts. To investigate whether the RNA that we found was due to the replicating virus, we assayed the respiratory tissues for sgRNA, which was not present in the donor lung tissue.
Although the detection of SARS-CoV-2 via culture-based techniques is considered the gold standard for determining viral infectivity, viral culture is labor intensive and requires biosafety level 3 facilities, which are not readily available in most clinical laboratories. Detecting viral RNA intermediates, such as sgRNA, could serve as a surrogate for viral replication. sgRNA consists of a negative sense RNA from the 3′ end of the viral genome joined with a leader segment derived from the 5′ end that is present in SARS-CoV-2 when the transcription is active, secondary to the discontinuous transcription events that are unique to coronaviruses.21, 22, 23, 24 Detecting SARS-CoV-2 sgRNA is accomplished using a simple 1-step reverse transcription PCR assay that can be easily performed in a clinical laboratory within 4 hours.
There is accumulating evidence that sgRNA assays can accurately predict viral infectivity. A recent prospective study evaluating the kinetics of sgRNA in 36 patients with COVID-19 found that the median time to negative sgRNA was 11 days from symptom onset, compared with 18 days for standard SARS-CoV-2 reverse transcription PCR (P < .001). Importantly, no virus was isolated by culture when cycle threshold values for sgRNA were >31.25 In another study comparing SARS-CoV-2 envelope sgRNA with viral culture, sgRNA detected replication-competent virus with a sensitivity of 97%, a positive predictive value of 94%, and a negative predictive value of 95%. There was strong concordance between the results of sgRNA and viral culture.21
A limitation of using sgRNA is that sgRNA transcripts are believed to become trapped in double-membrane or extracellular vesicles, and thus protected from cytoplasmic degradation,26 which might allow sgRNA transcripts to persist beyond the period of active viral replication. However, this would have a greater impact on the specificity of sgRNA for determining replication-competent virus, and not the sensitivity or negative predictive value of the assay, which is of greater significance in our scenario.21 , 24
On multiple occasions following lung transplantation, PCR for SARS-CoV-2 was negative on the recipient’s BAL samples, which is the most definitive clinical evidence against SARS-CoV-2 infection and transmission. This case builds upon earlier studies demonstrating the potential safety of using lung allografts from COVID-19–recovered donors12, 13, 14 and supports OPTN recommendations requiring a negative SARS-CoV-2 PCR from a donor LRT sample within 72 hours of lung procurement. However, when a donor’s history or timeline of illness is consistent with a resolved SARS-Co-V-2 infection but there is persistent SARS-CoV-2 positivity for an upper respiratory tract or LRT sample with genomic RNA testing, a negative sgRNA result would suggest against active viral replication and indicate a lower likelihood of SARS-CoV-2 transmission through the lung allograft. Because greater than two-thirds of Americans have had COVID-19, the persistence of genomic RNA in the donor organ tissue has the potential to occur frequently.
Although our findings are consistent with those of earlier reports describing successful lung transplantation utilizing lungs from donors with a prior SARS-CoV-2 infection, there are several key distinctions. First, SARS-CoV-2 RNA in the donor lung tissue was not detected by PCR, RNAscope, or sequencing in 2 earlier studies.16 , 18 Although SARS-CoV-2 RNA was detected by PCR in the donor lung tissue in a third report, it was unclear whether the RNA was in the tissue or within blood vessels in the tissue, and RNAscope was not performed to localize viral RNA transcripts to a single-cell level.17
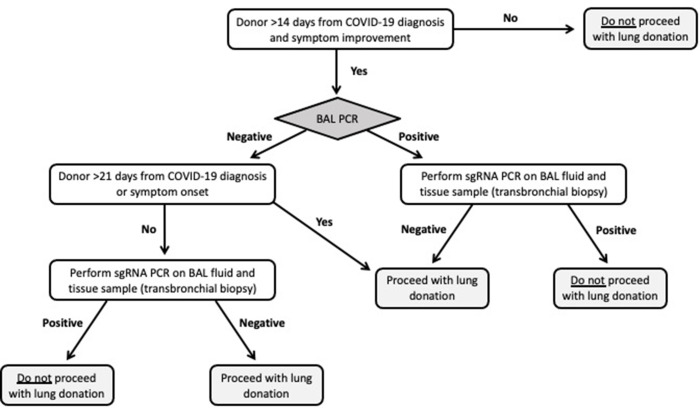
Given the high wait-list mortality of lung transplant recipients, it is impractical to exclude all donors with prior COVID-19 from lung allograft donation. However, it is imperative to develop a careful approach when considering donors with a history of COVID-19. Testing of LRT for SARS-CoV-2 RNA, as mandated by the United Network for Organ Sharing (UNOS)/OPTN policy,12 is a key component of donor assessment. Additionally, the time from symptom onset is another important consideration as the guidance from the UNOS/OPTN suggests that donor to recipient transmission is less likely to occur when the donor is between 21 and 90 days of symptom onset. SARS-CoV-2 proteins and RNA have, however, been identified in multiple organs, and data suggest that viral RNA can persist in tissues for prolonged periods of time following an infection.14 , 15 In a scenario in which a donor with a recently resolved COVID-19 infection has an LRT sample positive for SARS-CoV-2 by genomic RNA testing, sgRNA testing of an LRT sample and of a bronchial tissue via transbronchial biopsy could be performed to assess replication-competent viruses and guide organ utilization decisions (Fig. 3 ).
Fig. 3.
Potential algorithm incorporating subgenomic RNA (sg RNA) testing in prospective lung transplant donors with a recent SARS-CoV-2 infection. BAL, bronchoalveolar lavage; PCR, polymerase chain reaction.
Our case indicates that donor lungs containing SARS-CoV-2 genomic RNA can be transplanted with good outcomes despite the ongoing need for immunosuppression. Relatively low viral RNA copy numbers, no evidence of viral replication, and a normal lung architecture consistent with mild COVID-19 all likely contributed to the good outcome, in addition to the recipient’s immune response associated with recent recovery from SARS-CoV-2. The low viral copy numbers are consistent with a prolonged period following symptom onset.15 Because the date of COVID-19 symptom onset in the donor is unknown, we cannot comment on the time interval in the context of this case. Looking forward, the investigation of additional cases like this is needed to better understand how the persistence of viral genomic RNA in organs during transplantation impacts SARS-CoV-2 transmission and longer term allograft outcomes. Because genomic RNA detection cannot accurately predict the presence of replication-competent viruses and could result in discarding organs that do not pose the risk of viral transmission, further investigation of assays that can predict viral infectivity, such as sgRNA, is needed.
Funding
This work was supported by the intramural research programs of the NIH Clinical Center, National Institute of Allergy and Infectious Diseases, National Institute of Dental and Craniofacial Research, and National Cancer Institute.
Declaration of competing interest
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
These authors contributed equally as senior authors: Alison Grazioli and Daniel S. Chertow.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajt.2022.09.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shah M.B., Lynch R.J., El-Haddad H., Doby B., Brockmeier D., Goldberg D.S. Utilization of deceased donors during a pandemic: argument against using SARS-CoV-2-positive donors. Am J Transplant. 2020;20(7):1795–1799. doi: 10.1111/ajt.15969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kute V.B., Fleetwood V.A., Meshram H.S., Guenette A., Lentine K.L. Use of organs from SARS-CoV-2 infected donors: is it safe? A contemporary review. Curr Transplant Rep. 2021;8(4):281–292. doi: 10.1007/s40472-021-00343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kute V.B., Godara S., Guleria S., et al. Is it safe to be transplanted from living donors who recovered from COVID-19? Experience of 31 kidney transplants in a multicenter cohort study from India. Transplantation. 2021;105(4):842–850. doi: 10.1097/TP.0000000000003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koval C.E., Poggio E.D., Lin Y.C., Kerr H., Eltemamy M., Wee A. Early success transplanting kidneys from donors with new SARS-CoV-2 RNA positivity: a report of 10 cases. Am J Transplant. 2021;21(11):3743–3749. doi: 10.1111/ajt.16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romagnoli R., Gruttadauria S., Tisone G., et al. Liver transplantation from active COVID-19 donors: a lifesaving opportunity worth grasping? Am J Transplant. 2021;21(12):3919–3925. doi: 10.1111/ajt.16823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summary of current evidence and information-donor SARS-CoV-2 testing & organ recovery from donors with a history of COVID-19. Organ Procurement and Transplantation Network, US Department of Health and Human Services. Accessed January 25, 2022. https://optn.transplant.hrsa.gov/media/kkhnlwah/sars-cov-2-summary-of-evidence.pdf.
- 9.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar D., Humar A., Keshavjee S., Cypel M. A call to routinely test lower respiratory tract samples for SARS-CoV-2 in lung donors. Am J Transplant. 2021;21(7):2623–2624. doi: 10.1111/ajt.16576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaul D.R., Valesano A.L., Petrie J.G., et al. Donor to recipient transmission of SARS-CoV-2 by lung transplantation despite negative donor upper respiratory tract testing. Am J Transplant. 2021;21(8):2885–2889. doi: 10.1111/ajt.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notice of OPTN emergency policy change. Lower respiratory SARS-CoV-2 testing for lung donors. Organ Procurement and Transplantation Network, US Department of Health and Human Services. Accessed January 25, 2022. https://optn.transplant.hrsa.gov/media/4576/policy_notice_lunglowerrespiratorytesting_20210426.pdf.
- 13.Bhatnagar J., Gary J., Reagan-Steiner S., et al. Evidence of severe acute respiratory syndrome coronavirus 2 replication and tropism in the lungs, airways, and vascular endothelium of patients with fatal coronavirus disease 2019: an autopsy case series. J Infect Dis. 2021;223(5):752–764. doi: 10.1093/infdis/jiab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polak S.B., Van Gool I.C., Cohen D., von der Thüsen J.H., van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol. 2020;33(11):2128–2138. doi: 10.1038/s41379-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein S.R., Ramelli S.C., Grazioli A., et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy [published online ahead of print, 2022 Dec 14] Nature. 2022 doi: 10.1038/s41586-022-05542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Querrey M., Kurihara C., Manerikar A., et al. Lung donation following SARS-CoV-2 infection. Am J Transplant. 2021;21(12):4073–4078. doi: 10.1111/ajt.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceulemans L.J., Van Slambrouck J., De Leyn P., et al. Successful double-lung transplantation from a donor previously infected with SARS-CoV-2. Lancet Respir Med. 2021;9(3):315–318. doi: 10.1016/S2213-2600(20)30524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H.J., Shin D.H., Cho W.H., Kim D., Yeo H.J. Successful lung transplantation from a donor who had recovered from severe acute respiratory syndrome coronavirus 2 pneumonia. Ann Thorac Surg. 2022;113(5):e351–e354. doi: 10.1016/j.athoracsur.2021.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caniego-Casas T., Martínez-García L., Alonso-Riaño M., et al. RNA SARS-CoV-2 persistence in the lung of severe COVID-19 patients: a case series of autopsies. Front Microbiol. 2022;13:824967. doi: 10.3389/fmicb.2022.824967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K., Desai N.M., Resnick J., et al. Successful kidney transplantation from a deceased donor with severe COVID-19 respiratory illness with undetectable SARS-CoV-2 in donor kidney and aorta. Am J Transplant. 2022;22(5):1501–1503. doi: 10.1111/ajt.16956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos Bravo M., Berengua C., Marín P., et al. Viral culture confirmed SARS-CoV-2 subgenomic RNA value as a good surrogate marker of infectivity. J Clin Microbiol. 2022;60(1) doi: 10.1128/JCM.01609-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2(1):265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., et al. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18(6) doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J.Y., Bae J.Y., Bae S., et al. Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19. Clin Microbiol Infect. 2022;28(1):101–106. doi: 10.1016/j.cmi.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phuphuakrat A., Pasomsub E., Srichatrapimuk S., et al. Detectable duration of viable SARS-CoV-2, total and subgenomic SARS-CoV-2 RNA in noncritically ill COVID-19 patients: a prospective cohort study. Microbiol Spectr. 2022;10(3) doi: 10.1128/spectrum.00503-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandersen S., Chamings A., Bhatta T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat Commun. 2020;11(1):6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.