Extended Data Fig. 5. Comparison of SCAN2 and LiRA sSNV calls on human neurons.
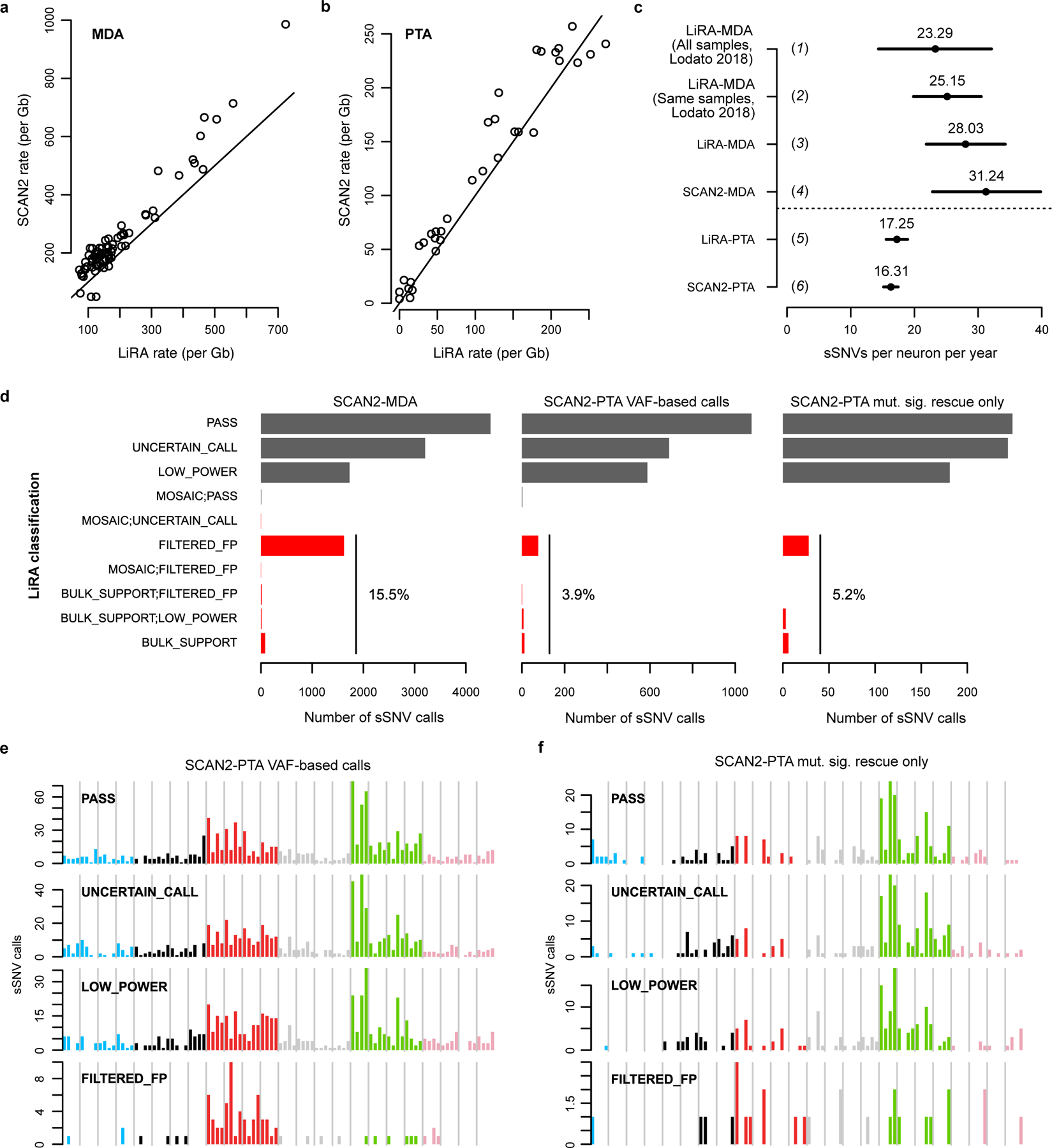
Single human neurons were previously analyzed by LiRA15, a specific but lower sensitivity approach for calling somatic SNVs. a-b. SCAN2 and LiRA extrapolations for the total (not called) sSNV burden per diploid Gb of human sequence from MDA- (a) and PTA-amplified (b) single neurons. Solid lines: y=x. c. Linear regression estimates for the number of sSNVs accumulated per neuron per year from several sources and analyses. Horizontal bars represent 95% C.I.s produced by confint applied to an lmer fit by the lme4 R package; centre points from fixef applied to the same fits. (1) LiRA rates taken from ref. 6, which used a larger set of n=91 MDA-amplified PFC neurons; (2) LiRA rates taken from ref. 6 using n=73 of the 75 MDA-amplified PFC neurons from subjects analyzed in this study (the two excluded neurons are 5087pfc-Rp3C5, an extreme outlier, and 4638-MDA-14); (3) rerun of LiRA on n=74 MDA-amplified neurons in (2) using the same input provided to SCAN2; (4) SCAN2 on n=74 MDA-amplified neurons; (5) LiRA on n=34 PTA-amplified neurons from donors also analyzed in ref. 6 (N.B. LiRA’s higher rate estimate in (c) occurs despite lower burden estimations in (b) due to differences in model intercepts: SCAN2 intercept=95.83, LiRA intercept=17.63); (6) SCAN2 on all n=52 PTA-amplified neurons generated here. d. LiRA classification of SCAN2 calls where reads linked to nearby germline heterozygous SNPs are available (black: likely true sSNVs, red: possible false positives). PASS is the highest quality LiRA class. UNCERTAIN and LOW_POWER indicate lack of linking reads to make a confident call, but no evidence of artifactual status is detected. All other classes (red) are interpreted as false positives. Percentages show the fraction of all false positive classes among SCAN2 calls. e-f. Raw mutation spectra for SCAN2 calls without (e) and with mutation signature-based calling (f) SCAN2 calls stratified by LiRA classification. The similarities between PASS and the two lower quality UNCERTAIN_CALL and LOW_POWER classes suggest that the majority of UNCERTAIN_CALL and LOW_POWER SCAN2 calls are true mutations. Confident false positives (FILTERED_FPs) possess a C>T dominated signature with lack of C>Ts at CpGs.
