Figure 5. Enrichment of neuronal mutations in functionally active genomic regions with tissue- and cell-type specificity.
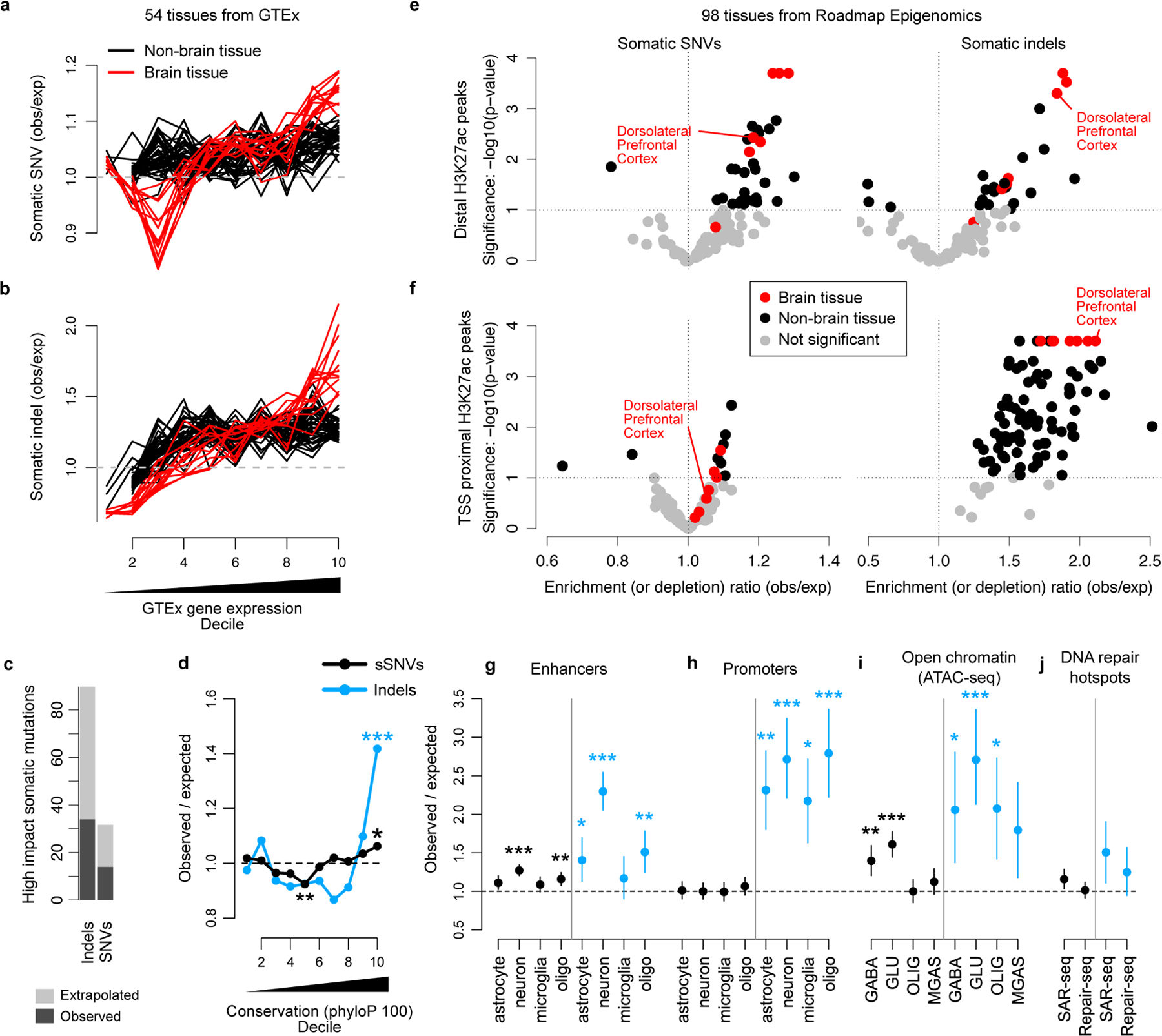
a-b. sSNV (a) and somatic indel (b) enrichment compared to local gene expression levels measured by GTEx. Each line corresponds to one GTEx tissue type; tissues from primary brain specimens are always shown in red. c. The number of high impact (classified HIGH by SnpEff; includes several severely protein altering effects such as stop gains, stop losses and frameshifts) sSNVs and somatic indels detected by SCAN2’s signature-based approach (dark grey) and extrapolation to autosomes (light grey). d. Mutation enrichment compared to local sequence conservation. e-f. Enrichment analysis of neuronal mutations in H3K27ac peaks from 98 Roadmap Epigenomics tissues. H3K27ac peaks are classified according to whether they are within 2 kb of an H3K4me3 peak in the same tissue (f, TSS proximal) or not (e, distal). Distal peaks are interpreted as intergenic enhancers. g-j. Mutation enrichment analysis of several datasets. Dorsolateral prefrontal cortex is shown since it most closely matches the neurons sequenced in this study. Cell-type specific enhancers (g) and promoters (h) from Nott et al. 2019; cell-type specific open chromatin regions (OCRs) measured by ATAC-seq from Hauberg et al. 2020 (i); DNA repair hotspots measured in induced human neurons (j) reported by Wu et al. 2021 (SAR-seq) and Reid et al. 2021 (Repair-seq). GABA, GABAergic neurons; GLU, glutamatergic neurons; OLIG, oligodendrocytes; MGAS, microglia and astrocytes. Error bars (g-j): 95% bootstrapping C.I. with n=104 bootstrap samplings; centre point: observed mutation count divided by the mean mutation count over bootstrap samplings. * - P < 0.01, ** - P < 0.001, *** P < 0.0001 by two-sided permutation test (Methods) without multiple hypothesis correction.
