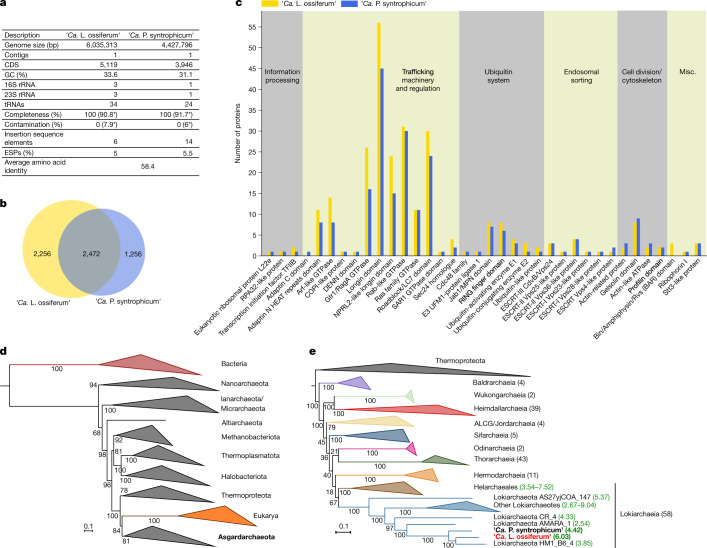
Fig. 2. Genome analysis and phylogenetic placement of ‘Ca. L. ossiferum’.
On the basis of the analysis of the closed genome of enrichment Loki-B35, we propose a description of the species ‘Ca. L. ossiferum’. a, The characteristics of the genome of ‘Ca. L. ossiferum’ in comparison to ‘Ca. P. syntrophicum’. Note the substantial difference in genome size. The values indicated by asterisks are the estimated values of contamination and completeness on the basis of the identification of marker genes performed by CheckM53 (Methods). b, The diagram shows to scale the number of shared clusters of orthologous proteins between ‘Ca. L. ossiferum’ and ‘Ca. P. syntrophicum’ as well as genome-specific clusters. A more detailed analysis is provided in Extended Data Fig. 4. c, A comparison of the occurrence of ESPs in ‘Ca. L. ossiferum’ and ‘Ca. P. syntrophicum’. Annotation of ESPs was performed according to the asCOG database2; general functional categories (on top) were added similar to earlier assignments1. Note that ‘Ca. L. ossiferum’ is enriched for ESPs of the following protein families associated with trafficking machineries: adaptin N heat repeats domain; Arf-like GTPase; Gtr1/RagA GTPase; longin domain; NPRL2-like longin domain. d,e, Maximum-likelihood (ML) phylogenies based on the concatenation of 23 universally conserved ribosomal proteins2. d, Tree of life showing Eukarya as a sister clade of Asgardarchaeota. e, ‘Ca. L. ossiferum’ and ‘Ca. P. syntrophicum’ belong to the same Lokiarchaeia class. Taxonomic assignments are based on the recently proposed classification11. Branch supports were calculated with 1,000 ultrafast bootstrap samples. The values in square brackets show the genome sizes of complete genomes (bold) and MAGs within Lokiarchaeia.