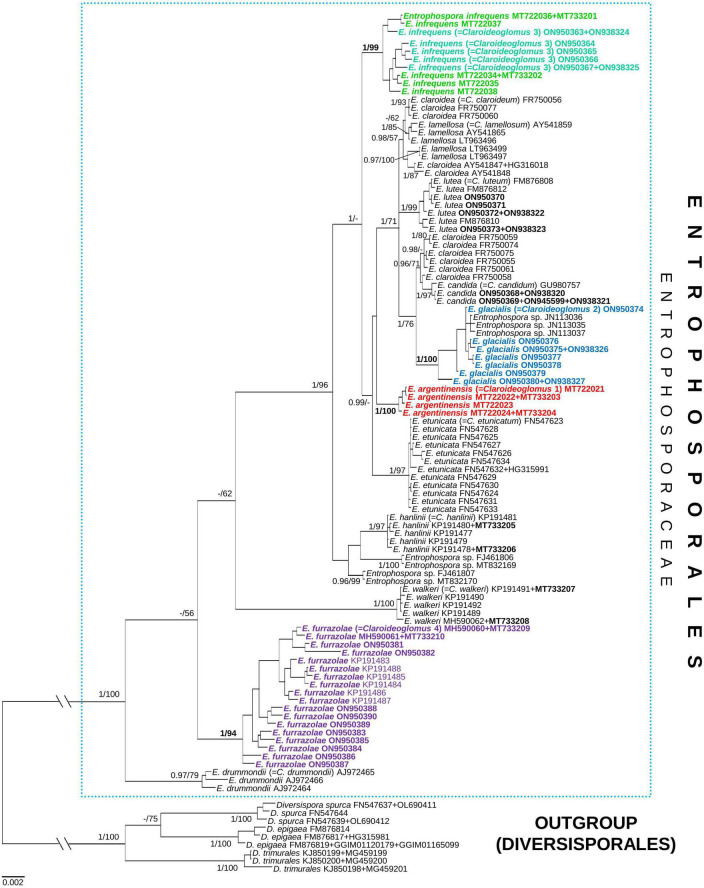
FIGURE 3.
A 50% majority-rule consensus tree from the Bayesian analysis of 45S nuc rDNA sequences concatenated with rpb1 sequences of Claroideoglomus 1, 2, 4 (newly described as Entrophospora argentinensis, E. glacialis, and E. furrazolae, respectively), Claroideoglomus 3 (a glomoid morph of the E. infrequens epitype), eight other species of Claroideoglomus sensu C. Walker and A. Schüßler, and three Diversispora species serving as outgroup. The former species names included in Entrophosporales are reported between brackets. The new species and the accession numbers of the sequences obtained in this study are in bold. The Bayesian posterior probabilities ≥0.90 and ML bootstrap values ≥50% are shown near the branches, respectively. Bar indicates 0.002 expected change per site per branch. The two basal branches were shortened to 20% in length to improve visibility (indicated by //).