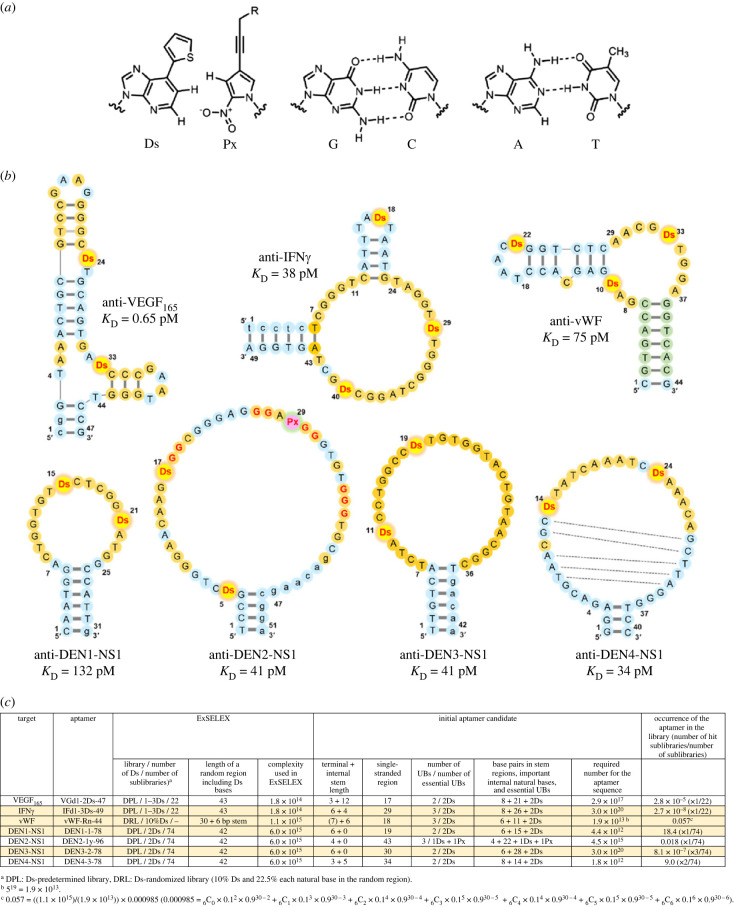
Figure 1.
High-affinity Ds-DNA aptamer candidates obtained by ExSELEX. (a) Chemical structures of a hydrophobic unnatural base pair, Ds–Px, and the natural A–T and G–C base pairs. (b) Predicted secondary structures of high-affinity Ds-DNA aptamers targeting VEGF165, IFNγ, von Willebrand factor A1-domain (vWF), and each serotype of dengue NS1 proteins. For the anti-DEN1/2/3/4-NS1 aptamers, their core parts are shown by removing the flanking primer regions. The bases in the randomized and primer regions are shown in large and small letters, respectively. The important bases (orange circles) were estimated from the doped selections (anti-VEGF165 and anti-IFNγ aptamers) or the obtained sequence motifs (anti-DEN1-, DEN2- and DEN4-NS1 aptamers). There is no information available for anti-vWF and DEN3-NS1, and thus the loop regions are tentatively estimated as the important bases. Green circles in the anti-vWF aptamer are the original constant complementary sequences for stem formation. In the anti-DEN2-NS1 aptamer, the four G tracts that form a G-quadruplex structure are shown by red Gs. The dashed lines in the anti-DEN4-NS1 aptamer represent a stem region found in the internal loop region. (c) Estimation of the occurrence of the aptamer candidates in the initial library based on ExSELEX data. (Online version in colour.)