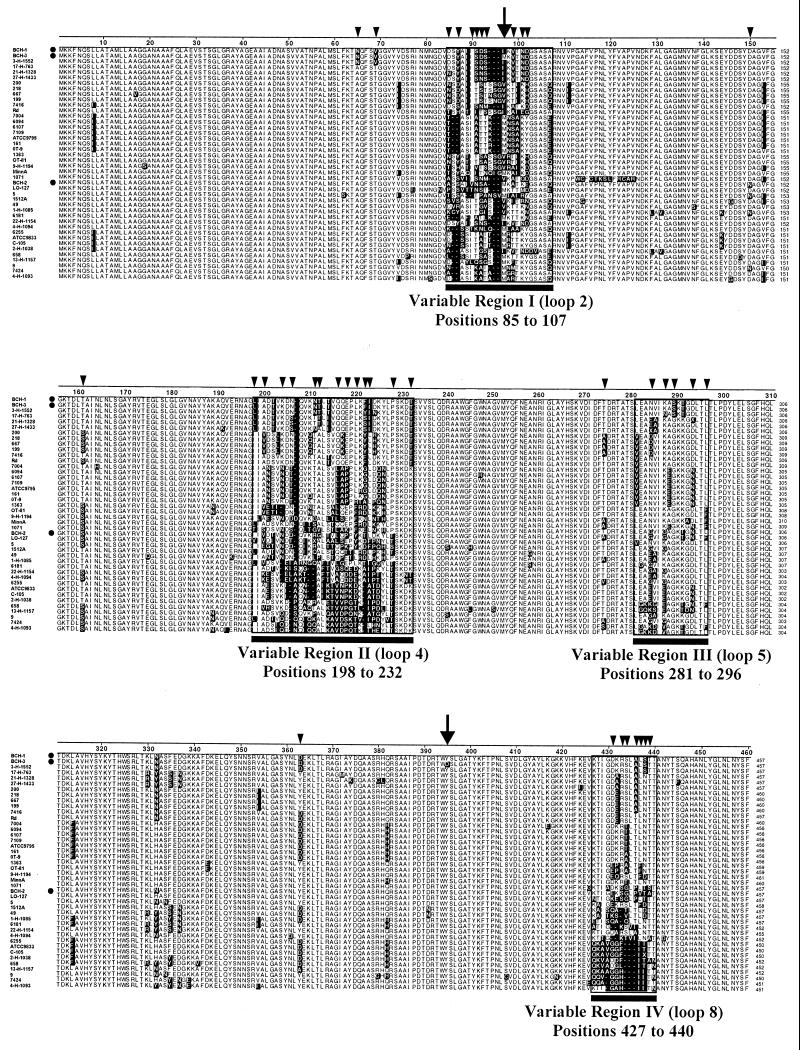
FIG. 1.
Alignment of deduced amino acid sequences of ompP1 alleles carried by the 42 phylogenically variant typeable H. influenzae and NTHI clinical isolates, listed as in Table 1. Dotted rows, isolates BCH-1, BCH-2, and BCH-3 used in the animal challenges; shaded residues, those that differ from the majority consensus sequence (not shown); black bars, variable regions. Numbers to the right of sequences indicate amino acid residue positions. Arrows above residues 97 (variable region I) and 395 (nonvariable region) identify the only two positions at which strains BCH-1 and BCH-3 differ. Smaller arrowheads indicate positions at which phylogenically variant strain BCH-2 differs from BCH-1, BCH-3, or both. Amino acid sequence alignment was performed with the LASERGENE software (DNASTAR), using the CLUSTAL multiple-alignment algorithm (30).