Figure 8.
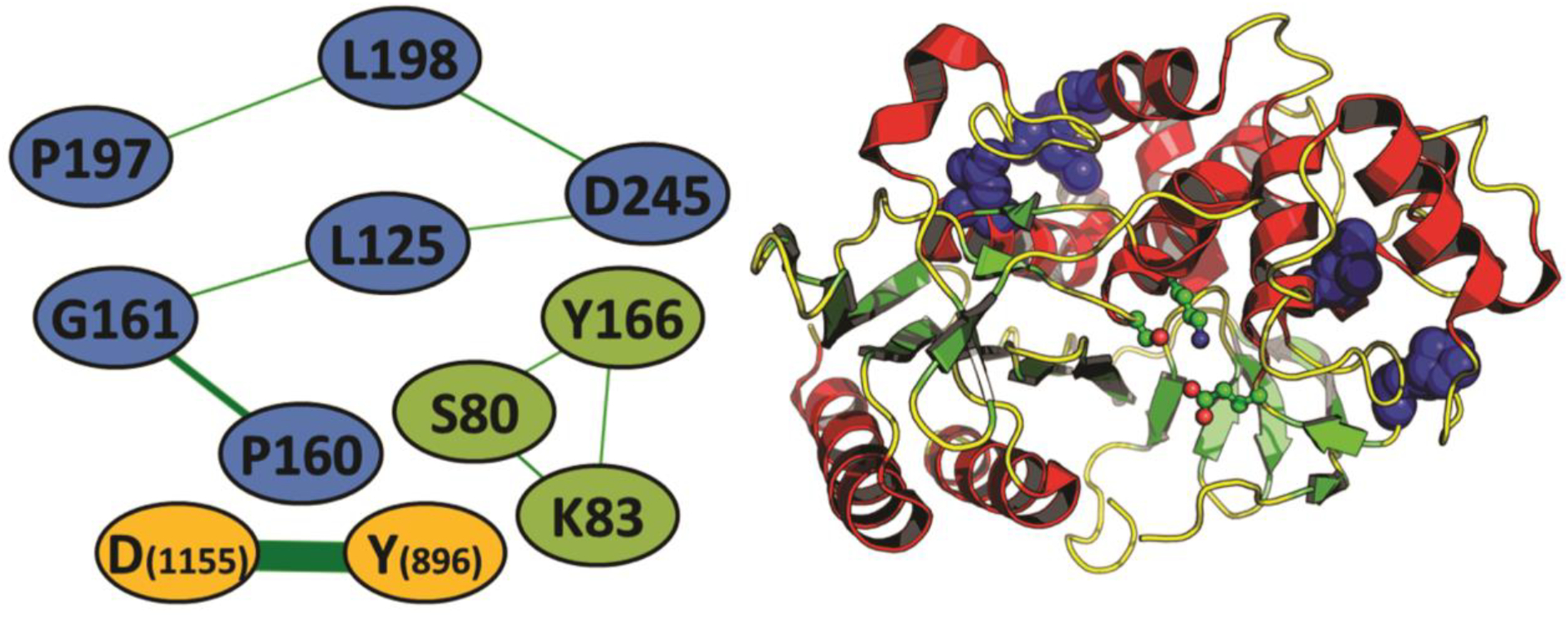
Coevolution network analyses of Lsa45 A) The co-evolution network in the β-lactamase family. Nodes are connected whenever there is coevolution (e.g., the presence of a proline in position 160 is usually followed by the presence of a glycine in position 161 and vice-versa). Nodes are colored according to the three sets of coevolving residues found in the analysis. E. coli AMPC is used, except for of the D1155-Y896 coevolving pair, which uses the alignment numbering due to its absence in both AMPC or Lsa45. B) location of the six-residue coevolving set (blue spheres) in the three-dimensional structure of a β-lactamase (PDB code: 1FCM) when compared to the three catalytic residues (as balls-and-sticks).
