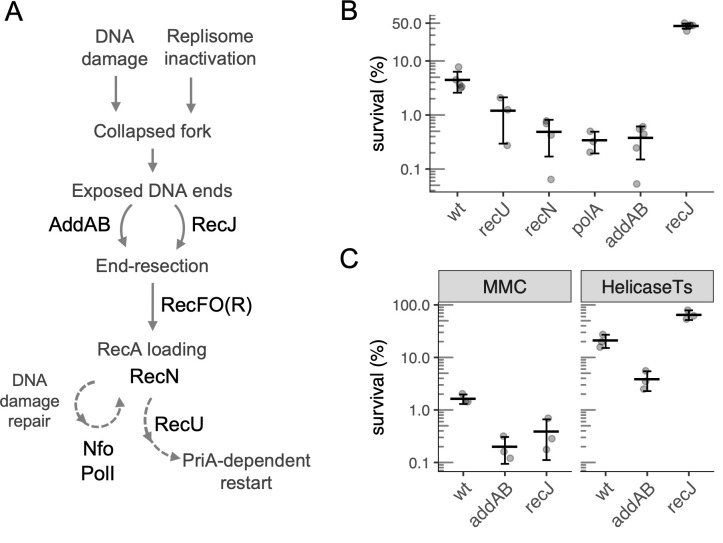
Fig 1. Pathways and genes involved in cell survival after arrest of DNA replication.
A. General view of the roles of the DNA repair and recombination genes identified in the Tn-seq screen. The dashed arrows represent multistep pathways. B. Percent survival of wild type (strain JMA222) and recU (GP891), polA (LSF41), recN (LSF298), addAB (LSF253) and recJ (LSF200) null mutants after 3 h of replication arrest caused by HPUra treatment. Percent survival is relative to cell viability at the time of arrest. All mutants were statistically different from wild type, as assessed by two-sample t-tests (P < 0.025). C. Effects of recJ and addAB null mutations on cell survival following replication arrest due to treatment with the DNA damaging agent mitomycin-C (MMC) or inactivation of the replicative helicase (DnaC). Left panel: survival of wild-type (JMA222), ΔaddAB (LSF253) and ΔrecJ (LSF200) cells after 3 h of treatment with 0.33 μg/ml of MMC. Right panel: wild-type, ΔrecJ and ΔaddAB cells carrying a dnaCts allele (LSF176, LSF274, and LSF326) were grown at permissive temperature (30°C) to early exponential phase and shifted to non-permissive temperature (49°C) for 4 h. Percent survival is relative to the population before treatment (left) or the temperature shift (right). The deletion mutants in both conditions were statistically different from wild type, as assessed by two-sample t-tests (P < 0.025). Throughout this work, error bars correspond to one standard deviation of the mean and each point is from an independent culture (biological replicates).