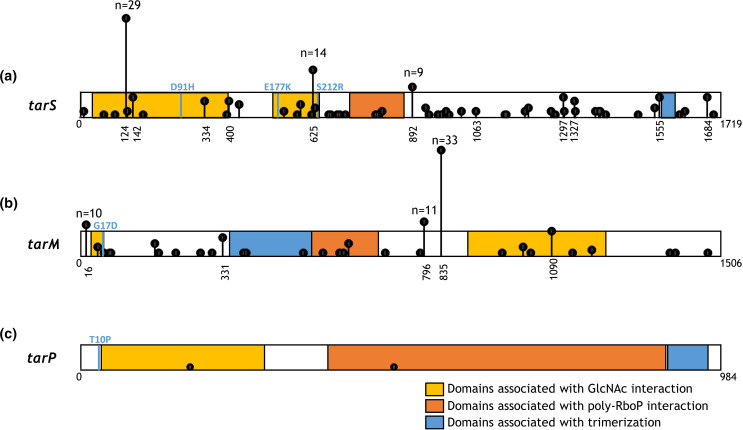
Fig. 4.
Tar-glycosyltransferase genes depicted as scaled-2D models with nucleotide position of premature stop codons. (a) Scaled 2D representation of tarS (nt 1719) containing two domains important for the GlcNAc interaction (nt 28–393 and 529–636; yellow), a poly-RboP interaction domain (nt 742–888; orange) and two residues associated with trimerization (nt 1561–1563 and 1594–1596; blue). Premature stop codons were identified in 165 isolates and are indicated by the vertical black lines that show position and frequency. For premature stop codons that were present in >4 isolates, the nucleotide position is shown, and for >8 the exact number of isolates is indicated. Amino acid substitutions are depicted in light blue. (b) tarM (1506 nt) contains two residues at the start (nt 49–54; yellow) and domain (nt 910–1233; yellow) that are important in the GlcNAc interaction. Domain 349–540 (blue) contains the HUB domain (formerly known as DUF1975) associated with TarM trimerization. Directly adjacent is the poly-RboP interaction domain (nt 543–699; orange). Premature stop codons (n=106 isolates) are indicated as in (a). (c) tarP (984 nt) contains a domain associated with the GlcNAc interaction (nt 31–285; yellow) followed by a poly-RboP interaction domain (385-789; orange). Domain 916–978 (blue) is associated with trimerization of the enzyme. Premature stop codons (n=2 isolates) are indicated as in (a).