Abstract
Women of African ancestry have the highest mortality from triple-negative breast cancer (TNBC) of all racial groups. To understand the genomic basis of breast cancer in the populations, we previously conducted genome-wide association studies and identified single nucleotide polymorphisms (SNPs) associated with breast cancer in Black women. In this study, we investigated the functional significance of the top associated SNP rs13074711. We found the SNP served as an enhancer variant and regulated TNFSF10 (TRAIL) expression in TNBC cells, with a significant association between the SNP genotype and TNFSF10 expression in breast tumors. Mechanistically, rs13074711 modulated the binding activity of c-MYB at the motif and thereby controlled TNFSF10 expression. Interestingly, TNFSF10 expression in many cancers was consistently lower in African Americans compared with European Americans. Furthermore, TNFSF10 expression in TNBC was significantly correlated with the expression of antiviral immune genes and was regulated by type I interferons (IFNs). Accordingly, loss of TNFSF10 resulted in a profound decrease in apoptosis of TNBC cells in response to type I IFNs and poly(I:C), a synthetic analogue of double stranded virus. Lastly, in a syngeneic mouse model of breast cancer, TNFSF10-deficiency in breast tumors decreased tumor-infiltrated CD4+ and CD8+ T cell quantities. Collectively, our results suggested that TNFSF10 plays an important role in the regulation of antiviral immune responses in TNBC, and the expression is in part regulated by a genetic variant associated with breast cancer in Black women. Our results underscore the important contributions of genetic variants to immune defense mechanisms.
Introduction
Black women of African Ancestry in the US experience the highest mortality rates from breast cancer despite having slightly lower incidences of the disease compared with women of other ethnic groups. There has been marked improvement in access to mammographic screening, which now exceeds or equals screening rates in non-Hispanic Whites (1). Although improved access has led to equal rates in the diagnosis of ductal carcinoma in situ, there has been no decline in the proportion of African Americans (AA) women diagnosed with triple-negative breast cancer (TNBC) of advanced stages (2,3). This indicates that, despite having access to proper screening and standard treatments (4), women of African descent are more likely to be diagnosed with aggressive types of breast cancer, have inferior therapeutic outcomes, poorer responsiveness to chemotherapy and more therapy-induced toxicity (3,5–8).
To understand the genomic basis of health disparities and to accelerate progress in the field, we conducted a large scale genome-wide association studies (GWAS) using combined samples of breast cancer cases and controls in Africa, Caribbean countries and the USA (4673 cases and 4774 controls of African ancestry in the discovery phase and 1984 cases and 2939 controls in the validation phase). We identified single nucleotide polymorphisms (SNPs) associated with estrogen receptor (ER)-negative breast cancer in women of African descent (5). The top SNP, rs13074711, is located at 3q26, 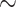 26.5 kb upstream of the TNFSF10 gene. Intriguingly, this region includes enhancer marks as well as an open chromatin structure, suggesting a potential role for the SNP (and linked SNPs) to serve as an enhancer that regulates TNFSF10 gene expression.
26.5 kb upstream of the TNFSF10 gene. Intriguingly, this region includes enhancer marks as well as an open chromatin structure, suggesting a potential role for the SNP (and linked SNPs) to serve as an enhancer that regulates TNFSF10 gene expression.
TNFSF10 (also well-known as TRAIL), a member of the tumor necrosis factor (TNF) superfamily, is a homotrimeric type II transmembrane ligand that preferentially induces apoptosis in tumor cells or transformed cells, but is not toxic to normal cells (6,7). Interaction of TNFSF10 with its receptors recruits the adaptor molecule FADD, activates caspases and degrades cellular components, ultimately leading to apoptosis of cancer cells. Interestingly, recent evidence suggests a significant role for TNFSF10/TRAIL in regulating antitumor immunity in cancer cells and the tumor microenvironment (TME) (8). TRAIL promotes granzyme B expression in cytotoxic T-cells, yet can also stimulate proliferation of regulatory T cells and M2 macrophages. These data suggest that TNFSF10 not only promotes cell death but also regulates immune cell function and proliferation.
Tumor immunity and immunotherapy have become increasingly important in treatment strategies for a variety of malignancies including advanced TNBC (9,10). The efficacy of cancer immunotherapy appears to depend on the host immune system recognizing and eliminating cancer cells (11). Increasing evidence supports a positive correlation between the presence of host antitumor immune responses and favorable patient outcomes for many cancers (12–15). As an example, tumors with a high density of tumor-infiltrating lymphoid cells (TILs) in the TME are more likely to respond to immune checkpoint inhibitors, whereas those with low or no TILs are less likely to respond to the inhibitors (16–18). Thus, interventions that promote antitumor immunity and render non-responding tumors into responding tumors bear tremendous therapeutic potential. Accordingly, cumulative evidence suggests that antiviral innate immune responses hold intrinsic anticancer benefits by promoting antitumor immunity and thereby increasing efficacy of chemotherapy and immunotherapy (11,19–23).
In this study, we discovered that the top associated SNP rs13074711 regulates TNFSF10 expression in TNBC cells, through the modulation of c-MYB binding activity at the motif. Furthermore, TNFSF10/TRAIL plays a significant role in regulating antiviral and antitumor immunity in TNBC. Our data suggest that genetic variants that drive aberrant expression of TNFSF10 may contribute to antitumor defense mechanism and immunotherapy responses in aggressive breast cancer, especially in women of African descent.
Results
Genotype-specific expression of TNFSF10 in TNBC cells from Black women
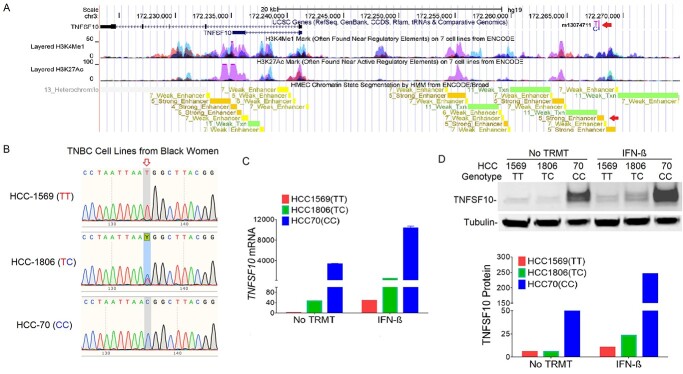
The top associated SNP rs13074711 is located 26.5 kb upstream of TNFSF10 (TRAIL) in the region of high histone H3 lysine 4 monomethylation (H3K4Me1) and histone H3 lysine 27 acetylation (H3K27Ac) modifications (Fig. 1A). Strong enhancer features of the SNP were also found in human mammary epithelial cells (HMECs), suggesting that the association between variants in the 3q26.21 region and breast cancer risk may be mediated by regulating TNFSF10 gene expression through the enhancer activities.
Figure 1.
Genotype-specific expression of TNFSF10 in TNBC cell lines from Black women. (A) The UCSC Genome Browser shows the locations of rs13074711 and TNFSF10, along with ENCODE histone modification marks for H3K4Me1 and H3K27Ac of seven cell types and enhancer marks in HMEC. Arrows indicate the location of rs13074711 and a strong enhancer mark in HMEC. (B) DNA sequencing chromatograms are shown in three TNBC cell lines (HCC1569, HCC1806 and HCC70) from Black women. The arrow indicates the position of rs31074711 and Y represents TC heterozygote. (C) Expression of TNFSF10 mRNA was measured by qRT-PCR in three cell lines with or without IFN-β treatment (24 h). Results were normalized to RNA18S and fold induction is shown relative to untreated HCC1569 cells. (D) Representative images (top) and a summary graph (bottom) of western blot analysis of TNFSF10 protein expression in three cells with or without IFN-β treatment. Tublin was used as an endogenous loading control. All data represent the mean and SD of n = 3–4 biological replicates and are representative of at least two independent experiments.
To evaluate the role of SNP rs13074711 in regulating TNFSF10 expression, we genotyped TNBC cell lines from Black women (Fig. 1B) and tested the correlation of rs13074711 genotypes with TNFSF10 expression (Fig. 1C and D). HCC-1569 cells with the TT genotype showed the lowest levels of mRNA (Fig. 1C) and protein expression (Fig. 1D), whereas those with the CC genotype showed the highest levels. Cells with the heterozygous genotype (TC) exhibited an expression level in-between homozygous (TT or CC)-genotype cells. Furthermore, the increased TNFSF10 expression in response to interferon (IFN)-β treatment is also genotype-dependent (the highest in CC-genotype cells and the lowest in TT-genotype cells). The results suggest that the SNP rs13074711 regulates TNFSF10 expression in TNBC cells from Black women.
CRISPR-Cas9 editing of rs13074711 alters TNFSF10 expression and IFN-β-induced apoptosis
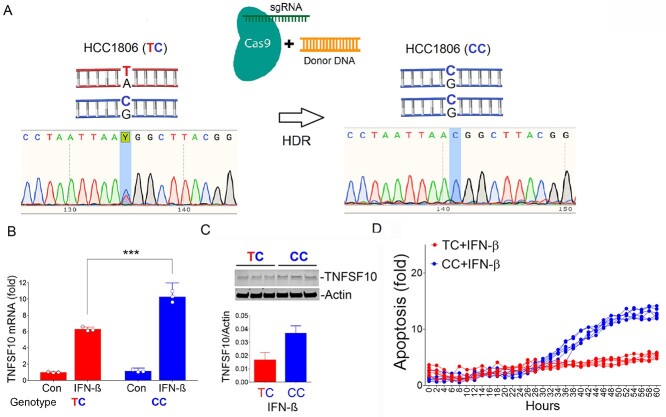
To directly test if rs13074711 genotypes have a significant effect on TNFSF10 expression, we engineered the TC genotype to the CC genotype in HCC-1806 cells, using CRISPR-Cas9 genome editing tools (Fig. 2A). An allelic change from TC to CC increased mRNA (Fig. 2B) and protein expression (Fig. 2C) of TNFSF10 in HCC-1806 cells, especially in response to IFN-β treatment. Furthermore, IFN-β-derived apoptosis was significantly greater in cells with the CC genotype compared with those with the TC genotype at 60-h post-treatment (12.8 ± 0.8 vs. 5.4 ± 0.4-fold increase) (Fig. 2D). Together, the data clearly showed that an allelic change in rs13074711 alters TNFSF10 expression and thereby affects IFN-β-induced apoptosis. In the expression quantitative locus (eQTL) analysis of breast tumors using data from The Cancer Genome Atlas (TCGA), we found that rs13074711 genotype was associated with TNFSF10 expression (P = 0.01) but not with other immune-related genes on different chromosomes (e.g. TNF, TLR3, IL8, DDX58, IFIH and OAS1), supporting our premise of cis-regulation of TNFSF10 expression by the SNP genotype.
Figure 2.
Regulation of TNFSF10 expression by rs31074711 Alleles. (A) Depicted is CRISPR-Cas9 genome editing of rs13074711 from TC genotype to CC genotype through HDR. DNA sequencing chromatograms showed the shift from T to C, after genome editing in HCC1806 cells. (B) qRT-PCR analysis of TNFSF10 mRNA in HCC1806 cells with TC or CC genotype of rs31074711 that were treated with IFN-β for 1 day. Results were normalized to RNA18S and fold induction is shown relative to the untreated control with TC genotype. (C) Western blot analysis of TNFSF10 protein expression in HCC1806 cells with TC or CC genotype 1-day post IFN-β treatment. Tublin was used as an endogenous loading control. (D) HCC1806 cells with TC or CC genotypes were treated with IFN-β and subjected to the IncuCyte Live-Cell Imaging System. Apoptosis was quantified using green fluorescent signals from caspase-3/7–positive apoptotic cells normalized to cell density. All data represent the mean and SD of n = 3–6 biological replicates and are representative of at least two independent experiments. ***P < 0.001.
rs13074711 regulates c-MYB binding activity in the TNFSF10 enhancer
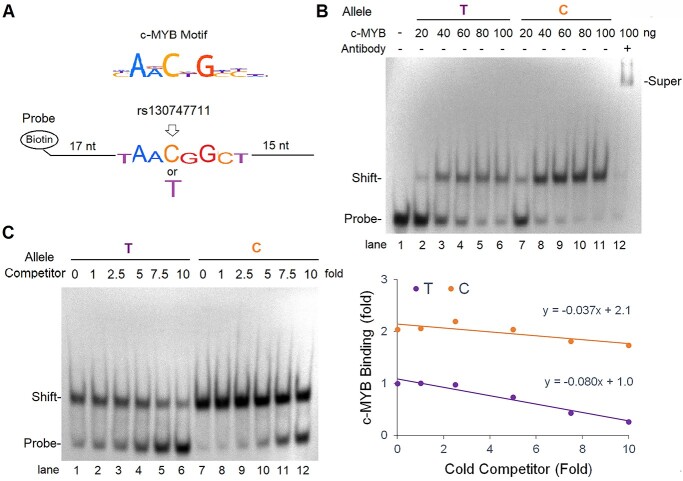
To determine if rs13074711 controls TNFSF10 expression through the regulation of enhancer activity, we conducted in silico analyses of cis-regulatory elements (motifs) of transcription factors. Our analysis predicts that an allelic change from T to C in rs13074711 may change the binding activity of c-MYB transcription factor at the motif (top, Fig. 3A). c-MYB is known to facilitate histone acetylation at super enhancer regions (24,25) and control cell proliferation, differentiation and apoptosis in many cell types (26). We thus examined whether rs13074711 regulates the enhancer activity of TNFSF10 through the c-MYB transcription factor, by conducting electrophoretic mobility shift assay (EMSA) using biotin-labeled oligonucleotides with the T or C allele (bottom, Fig. 3A). A stronger association of c-MYB was observed at the C allele of rs13074711 (lane 7–11, Fig. 3B) compared with the T allele (lane 2 to 6, Fig. 3B), for all concentrations (20–100 ng) of c-MYB protein treatments. The specificity of c-MYB’s interaction with the motif was confirmed by further retardation in mobility (super-shift), resulting from an interaction with anti-c-MYB antibodies (lane 12, Fig. 3B). Furthermore, when we added increasing amounts of cold competitors (non-biotin-labeled oligonucleotides, 1- to 10-fold of biotin-labeled probes) to the reaction, the strength of this interaction decreased in a dose-dependent manner because of its competition with non-biotin-labeled oligonucleotides (left, Fig. 3C). Notably, the decrease in the interaction was more rapid in the T allele compared with the C allele (slope − 0.080 vs. − 0.037) (right, Fig. 3C). These data indicate that the C allele of rs13074711 strengthens the binding site for c-MYB transcription factor at the TNFSF10 enhancer and thereby enhances TNFSF10 expression (Fig. 2).
Figure 3.
Regulation of c-MYB binding activity by rs13074711. (A) Depiction shows the c-MYB motif (top) and biotin-labeled oligonucleotides containing the cis-acting regulatory c-MYB element of the enhancer region (bottom). (B) EMSA was performed with biotin-labeled oligonucleotides (20 fmol) and recombinant c-MYB proteins (20–100 ng) with/without anti-MYB antibodies (200 ng). The mobilities of free oligonucleotides (probe), oligonucleotides associated with c-MYB (shift) or oligonucleotides associated both with c-MYB and anti-MYB antibodies (super-shift) were indicated. (C) Non-biotin-labeled oligonucleotides (cold competitor, 1–10-fold of hot probes) were incubated with c-MYB proteins (100 ng) before incubation with biotin-labeled probes (left). The densities of shift bands were plotted against the amounts of cold competitors added (right). Data are representative of at least three independent experiments.
TNFSF10 expression in breast tumors and other tumors
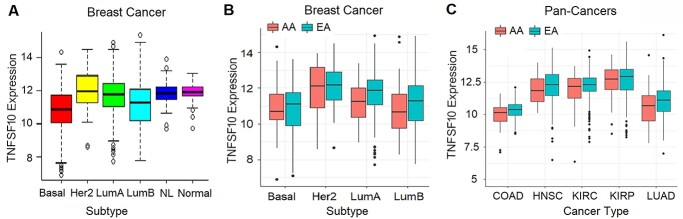
We next investigated TNFSF10 expression in patient samples using TCGA pan-cancer datasets (Fig. 4). When we analyzed the expression in breast tumors according to the molecular subtypes of breast cancer, we observed a strong association of the subtype with TNFSF10 expression in the multivariable model (P < 2.2 × 10−16) (Fig. 4A). Notably, the basal-like subtype had the lowest TNFSF10 expression among all subtypes, followed by the luminal B and luminal A subtypes.
Figure 4.
Comparison of TNFSF10 expression between EA and AA in breast and other tumors. TNFSF10 expression in the pan-cancer RNA-seq dataset was analyzed by (A) molecular subtypes of breast tumors (n = 1060); (B) by ethnicity and molecular subtypes of breast tumors; and (C) by ethnicity in five different types of tumors. The values are presented as log2 FPKM. Basal, basal-like; Her2, Her2-enriched, LumA, luminal A; LumB; NL, normal-like subtype of breast cancer. Normal indicates normal breast tissues. COAD, colon adenocarcinoma (n = 272); HNSC, head and neck squamous cell carcinoma (n = 492); KIRC, kidney renal clear cell carcinoma (n = 518); KIRP, kidney renal papillary cell carcinoma (n = 267); LUAD, lung adenocarcinoma (n = 440).
Next, we examined the association of TNFSF10 expression with ethnicity, after adjusting the subtype and age in the multiple linear regression. Race was modestly but significantly associated with TNFSF10 expression (P = 0.03). On average, TNFSF10 expression in breast cancer was 25% lower (0.4 unit in log2 scale) in AA compared with EA. Ethnic comparisons in each subtype of breast cancer showed that TNFSF10 expression was significantly lower in AA than EA in the luminal A subtype, with a general trend of lower expression in AA in the basal-like and luminal B subtypes (Fig. 4B).
For other tumors, we focused on six cancer types, which included at least 40 AA patient samples in TCGA pan-cancer dataset (Fig. 4C). TNFSF10 expression is ubiquitously lower in AA patients compared with EA patients in five types of cancers, with statistically significantly lower expression in AA in two types of cancers (colon and kidney cancers). Collectively, our data suggested reduced expression of TNFSF10 in AA patients, which may contribute to resistance to TNFSF10-driven apoptosis of cancer cells and poor clinical outcomes.
Correlation of TNFSF10 expression with antiviral immune genes in TNBC tumors
To identify genes and pathways co-regulated with TNFSF10, we next conducted genome-wide correlation analyses in breast cancer using TCGA datasets. TNFSF10 expression was significantly correlated with expression of 782 genes in basal-like breast cancer (P < 2.51 × 10−6) (Supplementary Material, Table S1), including significant enrichment of genes in immunogenic pathways and innate antiviral immune responses (Supplementary Material, Table S2). Examination of the correlated genes involved in innate immunity pathways revealed associations with known families of antiviral immunity genes, such as the RIG-I-like receptor signaling family and Toll-like receptor (TLR) signaling genes (Fig. 5A) (Supplementary Material, Fig. S1). These genes play important roles in stimulating type 1 IFN production and recognizing viral dsRNA to provide resistance to most RNA viruses. Other antiviral gene families identified from the GO term analysis included the tripartite motif family involved in viral infection and replication restriction, interferon-stimulating gene family, IFN-induced proteins with tetratricopeptide repeats (IFIT) and the oligoadenylate synthase (OAS) family, which is involved in viral RNA degradation.
Figure 5.
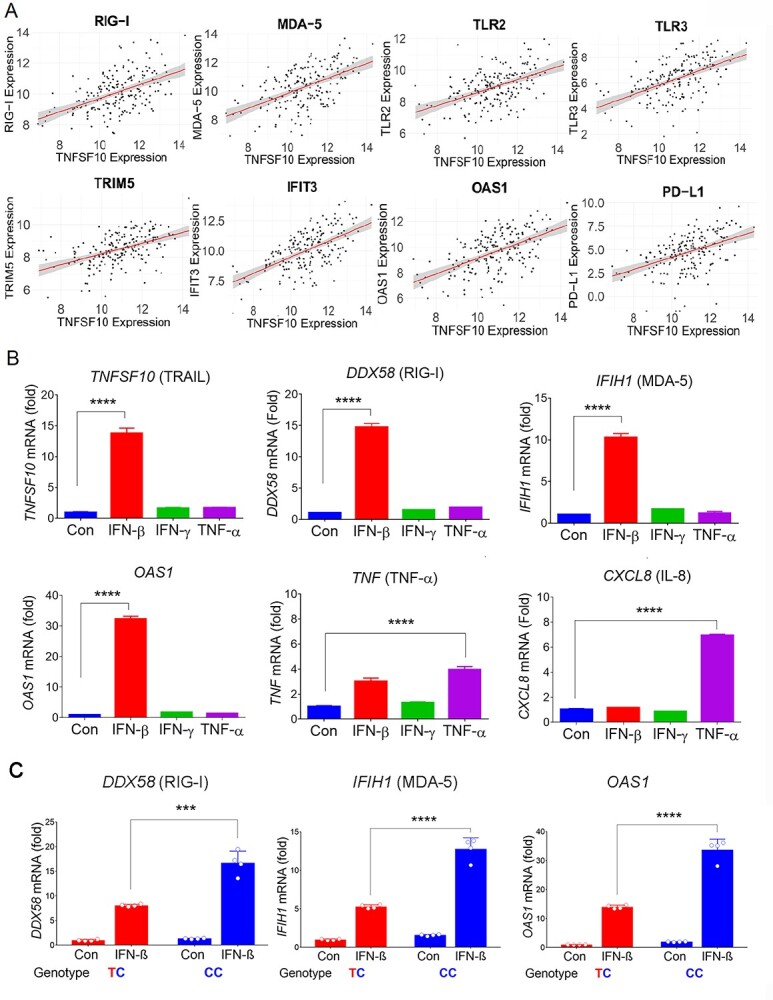
Correlation of TNFSF10 expression with anti-viral gene expression and regulation by cytokines. (A) Examples of key immune genes significantly correlated with TNFSF10 expression. Normalized RNA-seq data from TCGA-BRCA (n = 1076) were analyzed for correlation. The values are presented as log2 FPKM. (B) qRT-PCR analyses of TNFSF10, DDX58, IFIH1, OAS1, TNF, IL mRNA in HCC1806 cells that were treated with IFN-β, IFN-γ, or TNF-α for 1 day. (C) qRT-PCR analyses of DDX58, IFIH1, OAS1 mRNA in HCC1806 cells with TC or CC genotype of rs31074711 that were treated with IFN-β for 1 day. Results were normalized to RNA18S and fold induction is shown relative to untreated control cells. Data represent the mean and SD of n = 3–4 biological replicates and are representative of at least two independent experiments. ***P < 0.001; ****P < 0.0001.
The pathway analysis also identified a high-fold enrichment of several other inflammatory and adaptive immunity pathways from the gene set that included cytokine signaling processes, various processes of T-cell chemotaxis and proliferation, as well as MHC Class I antigen processing and presentation (Supplementary Material, Fig. S2). Notably, TNFSF10 expression had an additional highly significant correlation with expression of CD274 (PD-L1) (Fig. 5A), a therapeutic target and predictive biomarker in cancer immunotherapy.
TNFSF10 expression is induced by type I IFN, not by TNF-α
Because TNFSF10 expression is correlated with expression of antiviral immune genes in TNBC tumors, we assessed whether TNFSF10 expression is regulated by type I IFNs in TNBC cells (Fig. 5B). IFN-β dramatically increased TNFSF10 expression in TNBC cells, along with expressions of other classical antiviral genes [DDX58 (RIG-I), IFIH1 (MDA-5) and OAS1]. In contrast, TNFSF10 expression was minimally induced by IFN-γ or TNF-α treatment, which significantly stimulated TNF (TNF-α) and CXCL8 (IL-8) expression. Collectively, the data indicated that, although TNFSF10 is a member of TNF superfamily, its expression is not regulated by TNF-α-mediated inflammatory signaling, but rather by type I IFN-mediated innate immune signaling.
Correlation of antiviral gene expression with SNP rs13074711 genotypes
We next examined the relationship between antiviral gene expression and SNP genotypes in TNBC cells (Fig. 5C). This is partly because TNFSF10 expression is correlated with expression of antiviral immune genes, induced by IFN-β, and is also regulated by SNP rs13074711. We utilized CRISPR-Cas9 edited HCC-1806 cells with the TC or CC genotypes of rs13074711, which were correlated with TNFSF10 expression (Fig. 2). An allelic change from TC to CC significantly increased mRNA expression of antiviral genes [DDX58 (RIG-I), IFIH1 (MDA-5) and OAS1], especially in response to IFN-β treatment. Expression of RIG-I (DDX58) and MDA-5 (IFIH1) was 2.07-fold and 2.44-fold higher, respectively, in cells with the CC genotype compared with those with the TC genotype after IFN-β treatment. The data indicate the importance of the SNP in regulating TNFSF10 expression, which may subsequently contribute to antiviral immune responses in TNBC cells.
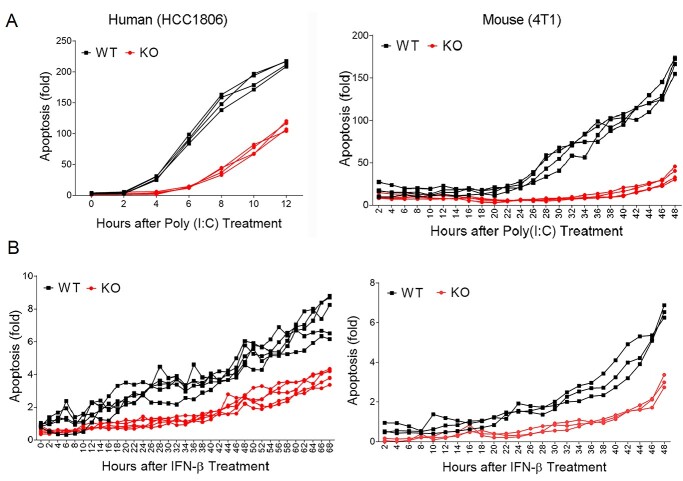
TNFSF10 plays an essential role in poly(I:C) or IFN-β-driven apoptosis
To investigate the role of TNFSF10 in regulating antiviral defense mechanism, we utilized CRISPR-Cas9 technology and deleted the TNFSF10 gene from TNBC cells (Supplementary Material, Fig. S3). Deletion of TNFSF10 did not change proliferation (Supplementary Material, Fig. S3C) or apoptosis (Supplementary Material, Fig. S3D), before stimulation of antiviral immune responses. However, when we treated cells with poly(I:C), a synthetic analogue of double-stranded RNA virus, we observed a clear difference in poly(I:C)-induced apoptosis between the wild-type (WT) and TNFSF10-KO cells from human (HCC1806) and mouse (4 T1) TNBC cell lines (Fig. 6A). Depletion of TNFSF10 clearly reduced poly (I:C)-induced apoptosis, while poly (I:C) treatment led to a dramatic increase in apoptosis of WT cells. Similarly, loss of TNFSF10 clearly reduced IFN-β-induced apoptosis, while IFN-β treatment significantly induced apoptosis of WT cells (Fig. 6B). Collectively, our data suggested that TNFSF10 plays an essential role in antiviral immunity-induced apoptosis in TNBC cells, partly through type I IFN signaling.
Figure 6.
Decreased apoptosis of TNFSF10-KO cells in response to poly(I:C) and IFN-β. (A and B) Apoptosis of TNFSF10-WT and KO cells was analyzed using the IncuCyte Live-Cell Imaging System after poly(I:C) (A) or IFN-β (B) treatment. Apoptosis was quantified using green fluorescent signals from caspase-3/7-positive apoptotic cells normalized to cell density. Data represent the mean and SD of n = 4–10 biological replicates and are representative of at least two independent experiments.
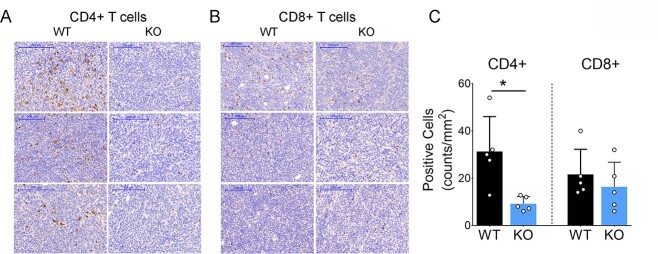
Loss of TNFSF10 inhibits T-cell infiltration into tumors, in vivo
Lastly, we investigated the effect of TNFSF10-deficient tumor cells in regulating immune cell infiltration and antitumor immunity, using an immunocompetent syngeneic mouse model of 4 T1 breast cancer (Fig. 7). Tumor cells with TNFSF10-KO or WT genotypes were engrafted into BALB/c mice, monitored for tumor growth for 27 days and harvested for immune staining at the termination of the mice. We did not observe significant differences in tumor growth between mice with TNFSF10-KO or WT tumors, consistent with our in vitro data showing no significant difference in proliferation between TNFSF10-KO and WT cells before stimulation of antiviral immune responses (Supplementary Material, Fig. S3C). However, there were clear differences in the levels of tumor-infiltrating CD4+ or CD8+ T cells between TNFSF10-KO and WT tumors (Fig. 7A and B), with adjacent lymph nodes used as positive controls for CD4 and CD8 staining (Supplementary Material, Fig. S4). The mean number of CD4+ cells in the WT group was 31.3 ± 6.6 (counts/mm2), which was dramatically reduced to 9.2 ± 1.3 in the knockout (KO) group (Fig. 7C). The average number of CD8+ cells was modestly reduced from 21.6 ± 4.7 in the WT group to 16.3 ± 4.6 in the KO group. These data together indicate an important role for TNFSF10 in regulating the TME and TILs, suggesting that reduction of TNFSF10 expression may contribute to an unfavorable TME and decreased sensitivity to chemotherapy and immunotherapy.
Figure 7.
Decreased infiltration of CD4+ T cells into TNFSF10-KO breast tumors, in vivo. (A and B) Representative images from IHC analyses of CD4+ or CD8+ T cells in 4 T1 breast tumors with TNFSF10-KO or WT genotypes engrafted into syngeneic mice. Tumor-infiltrating CD4+ or CD8+ T cells were stained with anti-mouse CD4 or anti-mouse CD8 antibodies (in brown) and compared between TNFSF10-KO and WT tumors. Original magnification of 200× was used for scoring. The scale bar of 200 μm was included in all images. (B) The density (number) of CD4 or CD8 positive cells was estimated by counting all immuno positive cells in five high-powered fields. The result was presented as a number of positively stained cells per 1 mm2 of each tumor (the mean and SD) from five tumors (**P < 0.01).
Discussion
Following our previous identification of genetic variants associated with ER-negative breast cancer in women of African descent (5), we investigated the molecular mechanisms of SNP influences on gene regulation and tumor progression in TNBC. The top SNP rs13074711 resided in the enhancer region of TNFSF10/TRAIL (Fig. 1) and regulated TNFSF10 expression in TNBC cells from Black women (Fig. 2). In silico analyses followed by experimental validation (EMSA) showed that rs13074711 regulated TNFSF10 expression through c-MYB activity at the enhancer (Fig. 3). Furthermore, comparison of TNFSF10 expression between AA and EA women revealed relatively lower expression of TNFSF10 in AA in breast tumors and other tumors (Fig. 4). Interestingly, TNFSF10 expression was significantly correlated with the expression of antiviral immune genes and was regulated by type I IFN, not by TNF, in TNBC cells (Fig. 5). Accordingly, CRISPR-Cas9 KO of TNFSF10 led to a profound decrease in apoptosis of TNBC cells in response to poly(I:C) and IFN-β (Fig. 6). Lastly, in vivo experiments showed that TNFSF10-deficient tumors reduced the levels of tumor-infiltrating CD4+ and CD8+ T cells (Fig. 7), and thereby regulated the TME and antitumor immunity. Collectively, our data showed that a genetic variant associated with breast cancer in women of African descent regulated TNFSF10 expression in TNBC, which plays a significant role in the regulation of antiviral and antitumor immune responses. Our results may help to understand the roles of genetic factors in the regulation of immune defense mechanism and to improve therapeutic options for TNBC patients, especially women of African ancestry.
It is worth emphasizing that our understanding of the genetic risk factors for human complex diseases remains limited, partly because of our incomplete understanding of the significance of non-coding regions in the human genome. The haploid human genome occupies a total of over 3 billion DNA base pairs, but only about 1.5% of the genome codes for proteins, far fewer than had been expected before its sequencing. The rest of the human genome consists of non-protein-coding sequences with unknown biologic functions. Because the majority of common risk alleles discovered through GWAS map to non-protein-coding regions, it has been challenging to determine the biological functionality of SNPs in the development of human diseases.
In this study, we applied CRISPR-Cas9 genome-editing tools to specifically edit rs13074711, which resides in a non-protein-coding region of the human genome and identified the biological role of rs31074711 in regulating TNFSF10 expression in breast cancer. We also conducted cis- and trans-eQTL analyses of breast tumors using data from TCGA, confirming a significant association of rs13074711 genotypes with TNFSF10 expression (P = 0.01) (through cis-eQTL analyses), but not with other immune-related genes on different chromosomes (trans-eQTL analyses). Considering that trans-eQTL identification is much more challenging than cis-eQTL because a greater number of SNP-gene pairs are tested for trans-association on different chromosomes (27), further studies with much larger sample sizes or new methodologies are warranted to test the SNP association with other genes on different chromosomes. Nevertheless, our study suggests that rs13074711 regulates c-MYB binding activity and thereby controls TNFSF10 expression. Although the signaling pathways of c-MYB in the regulation of TNFSF10 expression merit further investigation, our results revealed an unidentified role for the non-coding SNP rs31074711 in the regulation of gene expression and immune defense mechanism in TNBC cells.
Interestingly, despite the fact that TNFSF10 is one of the TNF superfamily members, its expression was induced by type I IFNs but not by TNF in TNBC cells. In support of this, previous findings showed the regulation of TNFSF10 expression by IFNα in blood cells (28,29). Furthermore, our data clearly showed that TNFSF10 expression was highly correlated with antiviral gene expression in TNBC tumors, rather than with inflammatory or apoptosis-related gene expression. More importantly, when we treated breast cancer cells with poly(I:C), a synthetic analogue of double-stranded RNA virus, depletion of TNFSF10 clearly reduced poly(I:C)-induced apoptosis, compared with WT cells which underwent severe apoptosis upon poly(I:C) treatment (Fig. 6). Our experimental results, along with TCGA data analyses, suggested that TNFSF10 may play a significant role in antiviral defense mechanisms in TNBC cells.
Although TRAIL’s capacity for inducing cancer cell death and clearance of virus infection has been investigated in depth (30,31), there is still limited knowledge regarding whether and to what extent TNFSF10/TRAIL signaling can affect the antiviral response and immune tumor environment in cancer cells, especially TNBC cells. This is partly because most mice models used for tumor xenografts were athymic nude mice or immune-deficient NOD/SCID mice (32,33), which were unable to produce most T cells and were unable to exhibit the actual TME. We thus utilized the immunocompetent 4 T1 syngeneic mouse model and determined the effects of TNFSF10-KO on regulation of T-cell infiltration in breast tumors. We found that tumor-infiltrating CD4+ T cells were significantly reduced in TNFSF10-KO tumors compared with the WT tumors, indicating that TNFSF10 could regulate immune cell infiltration of tumors and thereby TILs in the TME. Considering that tumors with a high density of TILs in the TME are more likely to respond to immune checkpoint inhibitors (16–18), our data suggest that TNBC tumors with dysregulated TNFSF10 expression might have a reduced sensitivity to immune checkpoint inhibitors, which warrants further investigation.
To our knowledge, this is the first functional study to identify genetic and molecular mechanism of SNPs associated with aggressive breast cancer in women of African ancestry. It is unfortunate that there remain huge gaps in our knowledge of the molecular pathways that lead to aggressive breast cancer in Black women, who have high prevalence of ER-negative breast cancer and the associated worse clinical outcomes. Our results revealed a novel role for the non-coding SNP rs31074711 in the regulation of TNFSF10 expression and immune defense responses in TNBC cells. We also identified the mechanism of rs31074711-regulated TNFSF10 expression, which involves the modulation of c-MYB binding activity at the motif. Furthermore, we identified consistently lower expression of TNFSF10 in many cancers of AA compared with EA patients. Lastly, our data from in vivo experiments demonstrated an important role for TNFSF10 in regulating immune cell infiltration of tumors in the TME of TNBC tumors. As increasing evidence shows that antiviral innate immune responses promote antitumor immunity and the efficacy of chemotherapy and immunotherapy (11,19–23), our results may increase understanding of breast cancer susceptibility genes that can influence cancer development/progression through the regulation of immune defense mechanisms. Better understanding of the roles of genetic variants in the regulation of tumor immune microenvironment may also improve therapeutic options for TNBC patients, especially women of African ancestry.
Materials and Methods
Animal study and handling
All animals were humanely handled and monitored for health conditions according to the Institutional Animal Care and Use Committee approved protocols. Eight-week-old female BALB/c mice (Charles River Laboratories, Wilmington, MA) were anesthetized via inhalation with 2% vaporized isoflurane and were injected with 1 × 105 4 T1 breast cancer cells (100 μl, 50% Matrigel) into the fourth inguinal mammary gland at the base of the nipple. Tumor measurements were performed weekly using calipers to calculate tumor volume using the formula: 1/2 (length × width2). The assessment was blind to the animal groups and performed consistently throughout the study. Mice were sacrificed at the end of the study (27 days after injection) and tumors were collected from animals in each group for downstream analysis.
Immunohistochemistry (IHC) of mouse tumors
Tumors collected from both study groups (n = 5, each) were fixed in 10% formalin for 24 h, dehydrated in 70% ethanol and embedded in paraffin. Five micrometer sections from each Formalin-Fixed Paraffin-Embedded (FFPE) block were used for H&E, anti-CD4 and anti-CD8 staining. The IHC staining was performed according to the standard protocol on Leica Bond RX automated Stainer. After deparaffinization and rehydration, tissue sections were treated with Target Retrieval Solution (Leica Biosystems, #AR9640) for heat treatment for 20 min. Anti-mouse CD4 antibody (Synaptic Systems, HistoSur #HS-360-117, Clone: 78H9D2, 1:200 dilution) and anti-mouse CD8 antibody (eBioscience Affymetrix, #14–0808-82, Clone: 4SM15, 1:800 dilution) were applied to tissue sections for 1-h incubation at room temperature. The antigen–antibody binding was detected by ImmPress anti-rat IgG Polymer-HRP kit (Vector Laboratories, #MP-7444) and DAB (Leica Biosystems, Bond Polymer Refine kit). The slides stained for CD4+ and CD8+ were digitized on a 3D HISTECH pannoramic whole slide scanner. The density (number) of CD4 or CD8-positive cells was estimated by counting all immune-positive cells in five high-powered fields (1 mm2 each). The scoring and calculation of an averaging score were conducted by a pathologist (GFK), as previously described (34,35). The result was presented as the number of positively stained cells per 1 mm2.
Cell culture
Breast cancer cell lines from humans (HCC1569, HCC1806 and HCC70) and mice (4 T1) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were authenticated for species and unique DNA profile using short tandem repeat analysis by the provider. Cells were cultured in media recommended by ATCC and tested for mycoplasma contamination using the MycoAlert kit (Lonza, #LT07–318).
CRISPR-Cas9 knockout (KO) and genome editing
The KO experiment was performed as previously described with modifications (36). Briefly, a pair of gRNAs (crRNAs and tracrRNAs) was designed to delete 16 693 bp of TNFSF10 sequence using Alt-R CRISPR-Cas9 guide RNA tools (Integrated DNA Technologies, IDT), and was synthesized by the company. After incubation with S.p.Cas9 Nuclease V3 (IDT, #1081058), the gRNAs were transfected into TNBC cells according to the manufacturer’s instructions. For genome editing, gRNAs were designed to target the TNFSF10 enhancer region containing SNP rs13074711, which were incubated with S.p.Cas9 Nuclease V3 and transfected into TNBC cells, along with Alt-R homology-directed repair (HDR) donor oligos (IDT) for homologous recombination. DNA sequences of gRNAs or donor oligos will be provided upon request.
qRT-PCR and western blot analyses
qRT-PCR was performed using the TaqMan Gene Expression Master Mix (Thermo Fisher Scientific, #4369016), along with TaqMan Assays for TNFSF10 (Thermo Fisher Scientific, Hs00921974_m1), DDX58 (IDT, Hs.PT.58.4273674), IFIH1 (IDT, Hs.PT.58.1224165), OAS1 (IDT, Hs.PT.58.2338899), TNF (IDT, Hs.PT.5845380900) or CXCL8 (IDT, Hs.PT.5839926886). The fold change in expression of each gene was calculated using the ΔΔCT method, with RNA18S (IDT, Hs.PT.39a.22214856.g) as an internal control. To detect protein expression of TNFSF10, we conducted western blot analyses as previously described (36), using anti-TNFSF10 (Cell Signaling, #3219, 1:300 dilution), anti-tubulin (Cell Signaling, #2128, 1:2000 dilution) and anti-β-actin (abcam, #ab8227, 1:10 000 dilution) antibodies. After incubating with a fluorescent secondary antibody (Li-Cor, #926–68 071, 1:20 000 dilution), the fluorescence signals were detected by Li-Cor Odyssey Classic Infrared Imaging System, according to the manufacturer’s protocol.
The IncuCyte live-cell proliferation and apoptosis analysis
Cells plated at 5000 cells/well for HCC1806 (and 4000 cells/well for 4 T1) in 96-well plates were transfected with poly(I:C)-LMW (InvivoGen, #tlrl-picw) using Lipofectamin 3000 (Thermo Fisher Scientific, #L3000015). We used 50 ng and 200 ng of poly(I:C)-LMW for HCC1806 and 4 T1 cells, respectively. For cytokine treatment, HCC1806 cells were treated with 100 units of recombinant human IFN-beta (IFN-β) 1a protein (R&D systems, #11415–1), whereas mouse 4 T1 cells were treated with 2 ng of recombinant mouse IFN-β protein (R&D Systems, #8234-MB-010). Images of live cells were acquired using a 10× objective every 2 h (4 images/well) with the IncuCyte® S3 Live-Cell Analysis System, as described previously (36).
EMSAs
The construct expressing c-MYB was generously provided by Dr Odd Gabrielsen (University of Oslo, Norway) and recombinant c-MYB proteins were purified following his protocol (37). The cis-acting regulatory element of c-MYB on the TNFSF10 enhancer was analyzed using ENCODE ChIP-seq data from the UCSC Genome Browser and the consensus sequences were identified by TF finder (http://tfbind.hgc.jp). On the basis of sequences, two biotin-labeled synthetic oligonucleotides were synthesized with the T or C allele (IDT) and annealed together. Twenty to 200 fmols of oligonucleotides were incubated with 20–200 ng of recombinant c-MYB proteins, 1–10 fmols of oligonucleotides or 100–200 ng of anti-c-MYB antibodies (Cell Signaling, #12319) in the presence of 2.5% glycerol, 5 mM MgCl2 and 0.05% NP-40 using the LightShift Chemiluminescent EMSA kit (ThermoFisher, #20148, as previously described (36).
TCGA data analysis
TCGA pan-cancer RNA-seq and clinical data were used to examine the distributions of TNFSF10 expression across different subtypes of TCGA breast invasive carcinoma (TCGA-BRCA) and across different cancer types. First, we analyzed ethnic differences in TNFSF10 expression in TCGA-BRCA, using linear regressions after adjusting the age at diagnosis. We also examined racial differences in TNFSF10 expression in six other common cancers with at least 40 AA patients. Second, we conducted a correlation analysis of gene expression in female breast cancer patients with the ‘Basal’ PAM50 subtype for TNFSF10 expression with all other genes (n = 19 929) using the Pearson correlation method. An empirical P < 2.51 × 10−6 was considered statistically significant after correcting for multiple testing by the Bonferroni method (0.05/19929). To identify enriched pathways and biological processes of genes significantly correlated to TNFSF10, pathway analysis of 782 identified genes was performed using the DAVID’s Functional Annotation Tool (https://david.ncifcrf.gov/). Gene Ontology (GO) terms and BioCarta pathways were used for pathway analysis and functional annotation. GO terms were further filtered for redundant and obsolete terms with the Revigo tool (http://revigo.irb.hr/) at medium (0.7) setting. We also conducted eQTL analysis using breast tumors from TCGA, as described previously (38). Briefly, we examined an association between the SNP rs13074711 with the expression of TNFSF10 (cis-eQTL) or other immune-related genes on different chromosomes (trans-eQTL). Age at diagnosis, molecular subtype, race and copy number change were adjusted for in the linear regression models.
Statistical analysis of experimental data
Each experiment was repeated at least three times. The mean ± standard deviation (SD) of TNFSF10 mRNA or protein expression is presented and compared between two groups using the unpaired t-test and between three or more groups using the one-way ANOVA. Statistical significance was set at *P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001. Plots were generated using Graphpad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA). Two-sided P values < 0.05 were considered statistically significance.
Supplementary Material
Acknowledgements
We appreciate Lise Sveen’s careful review on the manuscript.
Conflict of Interest statement. O.I.O. serves on the Advisory Boards for CancerIQ and Tempus. All other authors have no conflict of interest to declare.
Contributor Information
Yoo Jane Han, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Jing Zhang, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Ashley Hardeman, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Margaret Liu, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Olga Karginova, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Roger Romero, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Galina F Khramtsova, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Yonglan Zheng, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Dezheng Huo, Department of Public Health Sciences, University of Chicago, Chicago, IL 60637, USA.
Olufunmilayo I Olopade, Section of Hematology/Oncology & Center for Clinical Cancer Genetics, Department of Medicine, University of Chicago, Chicago, IL 60637, USA.
Funding
Breast Cancer Research Foundation BCRF-21-071 (to O.I.O. and D.H.), National Cancer Institute (NCI) P20CA233307 (to O.I.O. and D.H.), R01CA228198 (to D.H.) and R01CA242929 (to D.H.), NCI PA12-149 (to A.H.), and the Kiphart Global Health Research Fund (to O.I.O.). O.I.O. is an American Cancer Society Clinical Research Professor.
Authors’ Contributions
Conception and design: Y.J. Han, O. I. Olopade, D. Huo.
Development of methodology: Y. J. Han, J. Zhang, A. Hardeman, O. Karginova, R. Romero, G. F. Khramtsova.
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): O. I. Olopade.
Analysis and interpretation of data (e.g. statistical analysis, biostatistics, computational analysis): Y.J. Han, M. Liu, Y. Zheng, D. Huo.
Writing, review, and/or revision of the manuscript: Y.J. Han, M. Liu, Y. Zheng, D. Huo, O. I. Olopade.
Administrative, technical, or material support (i.e. reporting or organizing data, constructing databases): Y.J. Han, O. I. Olopade.
Study supervision: Y.J. Han, O. I. Olopade.
References
- 1. Carey, L.A., Perou, C.M., Livasy, C.A., Dressler, L.G., Cowan, D., Conway, K., Karaca, G., Troester, M.A., Tse, C.K., Edmiston, S. et al. (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA, 295, 2492–2502. [DOI] [PubMed] [Google Scholar]
- 2. Iqbal, J., Rochon, P. and Ginsburg, O. (2015) Racial and ethnic differences in breast cancer diagnosis and survival--reply. JAMA, 313, 1475–1476. [DOI] [PubMed] [Google Scholar]
- 3. Dietze, E.C., Sistrunk, C., Miranda-Carboni, G., O'Regan, R. and Seewaldt, V.L. (2015) Triple-negative breast cancer in African-American women: disparities versus biology. Nat. Rev. Cancer, 15, 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mortel, M., Rauscher, G.H., Murphy, A.M., Hoskins, K. and Warnecke, R.B. (2015) Racial and ethnic disparity in symptomatic breast cancer awareness despite a recent screen: the role of tumor biology and mammography facility characteristics. Cancer Epidemiol. Biomark. Prev., 24, 1599–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huo, D., Feng, Y., Haddad, S., Zheng, Y., Yao, S., Han, Y.J., Ogundiran, T.O., Adebamowo, C., Ojengbede, O., Falusi, A.G. et al. (2016) Genome-wide association studies in women of African ancestry identified 3q26.21 as a novel susceptibility locus for oestrogen receptor negative breast cancer. Hum. Mol. Genet., 25, 4835–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wiley, S.R., Schooley, K., Smolak, P.J., Din, W.S., Huang, C.P., Nicholl, J.K., Sutherland, G.R., Smith, T.D., Rauch, C., Smith, C.A. et al. (1995) Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity, 3, 673–682. [DOI] [PubMed] [Google Scholar]
- 7. Wang, S. and El-Deiry, W.S. (2003) TRAIL and apoptosis induction by TNF-family death receptors. Oncogene, 22, 8628–8633. [DOI] [PubMed] [Google Scholar]
- 8. Sag, D., Ayyildiz, Z.O., Gunalp, S. and Wingender, G. (2019) The Role of TRAIL/DRs in the Modulation of Immune Cells and Responses. Cancer, 11, 1469–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adams, S., Diamond, J.R., Hamilton, E., Pohlmann, P.R., Tolaney, S.M., Chang, C.W., Zhang, W., Iizuka, K., Foster, P.G., Molinero, L. et al. (2019) Atezolizumab plus nab-paclitaxel in the treatment of metastatic triple-negative breast cancer with 2-year survival follow-up: a phase 1b clinical trial. JAMA Oncol., 5, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmid, P., Adams, S., Rugo, H.S., Schneeweiss, A., Barrios, C.H., Iwata, H., Dieras, V., Hegg, R., Im, S.A., Shaw Wright, G. et al. (2018) Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med., 379, 2108–2121. [DOI] [PubMed] [Google Scholar]
- 11. Gujar, S., Pol, J.G., Kim, Y., Lee, P.W. and Kroemer, G. (2018) Antitumor benefits of antiviral immunity: an underappreciated aspect of oncolytic virotherapies. Trends Immunol., 39, 209–221. [DOI] [PubMed] [Google Scholar]
- 12. Pitt, J.M., Vetizou, M., Daillere, R., Roberti, M.P., Yamazaki, T., Routy, B., Lepage, P., Boneca, I.G., Chamaillard, M., Kroemer, G. et al. (2016) Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity, 44, 1255–1269. [DOI] [PubMed] [Google Scholar]
- 13. Kroemer, G., Senovilla, L., Galluzzi, L., Andre, F. and Zitvogel, L. (2015) Natural and therapy-induced immunosurveillance in breast cancer. Nat. Med., 21, 1128–1138. [DOI] [PubMed] [Google Scholar]
- 14. Chen, D.S. and Mellman, I. (2017) Elements of cancer immunity and the cancer-immune set point. Nature, 541, 321–330. [DOI] [PubMed] [Google Scholar]
- 15. Yang, Y. (2015) Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Investig., 125, 3335–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smyth, M.J., Ngiow, S.F., Ribas, A. and Teng, M.W. (2016) Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol., 13, 143–158. [DOI] [PubMed] [Google Scholar]
- 17. Fend, L., Yamazaki, T., Remy, C., Fahrner, C., Gantzer, M., Nourtier, V., Preville, X., Quemeneur, E., Kepp, O., Adam, J. et al. (2017) Immune checkpoint blockade, immunogenic chemotherapy or IFN-alpha blockade boost the local and abscopal effects of oncolytic virotherapy. Cancer Res., 77, 4146–4157. [DOI] [PubMed] [Google Scholar]
- 18. Nagarsheth, N., Wicha, M.S. and Zou, W. (2017) Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol., 17, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonaventura, P., Shekarian, T., Alcazer, V., Valladeau-Guilemond, J., Valsesia-Wittmann, S., Amigorena, S., Caux, C. and Depil, S. (2019) Cold tumors: a therapeutic challenge for immunotherapy. Front. Immunol., 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newman, J.H., Chesson, C.B., Herzog, N.L., Bommareddy, P.K., Aspromonte, S.M., Pepe, R., Estupinian, R., Aboelatta, M.M., Buddhadev, S., Tarabichi, S. et al. (2020) Intratumoral injection of the seasonal flu shot converts immunologically cold tumors to hot and serves as an immunotherapy for cancer. Proc. Natl. Acad. Sci. U. S. A., 117, 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pyeon, D., Vu, L., Giacobbi, N.S. and Westrich, J.A. (2020) The antiviral immune forces awaken in the cancer wars. PLoS Pathog., 16, e1008814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simoni, Y., Becht, E., Fehlings, M., Loh, C.Y., Koo, S.L., Teng, K.W.W., Yeong, J.P.S., Nahar, R., Zhang, T., Kared, H. et al. (2018) Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature, 557, 575–579. [DOI] [PubMed] [Google Scholar]
- 23. Zhao, H., Ning, S., Nolley, R., Scicinski, J., Oronsky, B., Knox, S.J. and Peehl, D.M. (2017) The immunomodulatory anticancer agent, RRx-001, induces an interferon response through epigenetic induction of viral mimicry. Clin. Epigenetics, 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuglerud, B.M., Ledsaak, M., Rogne, M., Eskeland, R. and Gabrielsen, O.S. (2018) The pioneer factor activity of c-Myb involves recruitment of p300 and induction of histone acetylation followed by acetylation-induced chromatin dissociation. Epigenetics Chromatin, 11, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mansour, M.R., Abraham, B.J., Anders, L., Berezovskaya, A., Gutierrez, A., Durbin, A.D., Etchin, J., Lawton, L., Sallan, S.E., Silverman, L.B. et al. (2014) Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science, 346, 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fry, E.A. and Inoue, K. (2019) c-MYB and DMTF1 in cancer. Cancer Investig., 37, 46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shan, N., Wang, Z. and Hou, L. (2019) Identification of trans-eQTLs using mediation analysis with multiple mediators. BMC Bioinformatics, 20, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tecchio, C., Huber, V., Scapini, P., Calzetti, F., Margotto, D., Todeschini, G., Pilla, L., Martinelli, G., Pizzolo, G., Rivoltini, L. et al. (2004) IFNalpha-stimulated neutrophils and monocytes release a soluble form of TNF-related apoptosis-inducing ligand (TRAIL/Apo-2 ligand) displaying apoptotic activity on leukemic cells. Blood, 103, 3837–3844. [DOI] [PubMed] [Google Scholar]
- 29. Kayagaki, N., Yamaguchi, N., Nakayama, M., Eto, H., Okumura, K. and Yagita, H. (1999) type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: a novel mechanism for the antitumor effects of type I IFNs. J. Exp. Med., 189, 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strater, J. and Moller, P. (2004) TRAIL and viral infection. Vitam. Horm., 67, 257–274. [DOI] [PubMed] [Google Scholar]
- 31. Ishikawa, E., Nakazawa, M., Yoshinari, M. and Minami, M. (2005) Role of tumor necrosis factor-related apoptosis-inducing ligand in immune response to influenza virus infection in mice. J. Virol., 79, 7658–7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu, W., Zhang, H., Shi, Y., Song, M., Zhu, B. and Wei, L. (2013) Oncolytic adenovirus encoding tumor necrosis factor-related apoptosis inducing ligand (TRAIL) inhibits the growth and metastasis of triple-negative breast cancer. Cancer Biol. Ther., 14, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greer, Y.E., Gilbert, S.F., Gril, B., Narwal, R., Peacock Brooks, D.L., Tice, D.A., Steeg, P.S. and Lipkowitz, S. (2019) MEDI3039, a novel highly potent tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptor 2 agonist, causes regression of orthotopic tumors and inhibits outgrowth of metastatic triple-negative breast cancer. Breast Cancer Res., 21, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stasikowska-Kanicka, O., Wagrowska-Danilewicz, M. and Danilewicz, M. (2018) Immunohistochemical analysis of Foxp3(+), CD4(+), CD8(+) Cell infiltrates and PD-L1 in oral squamous cell carcinoma. Pathol. Oncol. Res., 24, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee, H.J., Kim, Y.A., Sim, C.K., Heo, S.H., Song, I.H., Park, H.S., Park, S.Y., Bang, W.S., Park, I.A., Lee, M. et al. (2017) Expansion of tumor-infiltrating lymphocytes and their potential for application as adoptive cell transfer therapy in human breast cancer. Oncotarget, 8, 113345–113359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han, Y.J., Zhang, J., Lee, J.H., Mason, J.M., Karginova, O., Yoshimatsu, T.F., Hao, Q., Hurley, I., Brunet, L.P., Prat, A. et al. (2021) The BRCA1 pseudogene negatively regulates antitumor responses through inhibition of innate immune defense mechanisms. Cancer Res., 81, 1540–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bergholtz, S., Andersen, T.O., Andersson, K.B., Borrebaek, J., Luscher, B. and Gabrielsen, O.S. (2001) The highly conserved DNA-binding domains of A-, B- and c-Myb differ with respect to DNA-binding, phosphorylation and redox properties. Nucleic Acids Res., 29, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adedokun, B., Du, Z., Gao, G., Ahearn, T.U., Lunetta, K.L., Zirpoli, G., Figueroa, J., John, E.M., Bernstein, L., Zheng, W. et al. (2021) Cross-ancestry GWAS meta-analysis identifies six breast cancer loci in African and European ancestry women. Nat. Commun., 12, 4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.