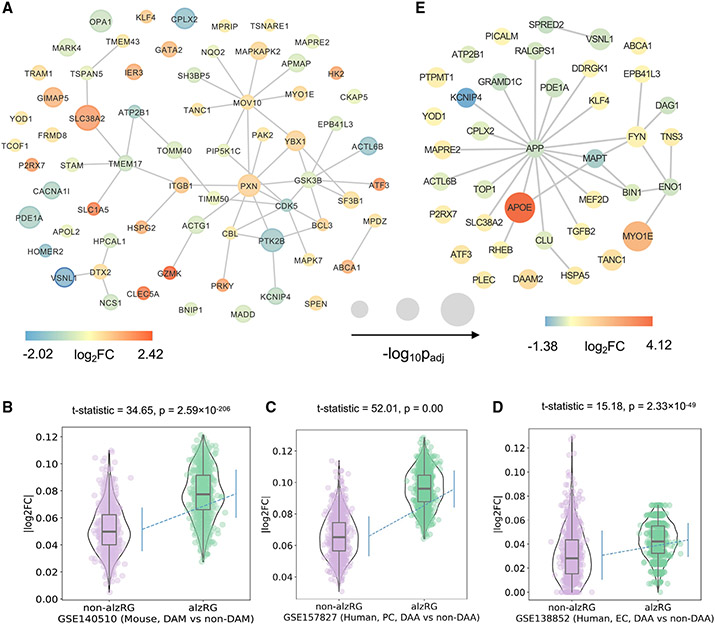
Figure 4. Transcriptomics-based observation of 156 prioritized AD-risk genes (alzRGs) by NETTAG.
(A) Visualization of 67 predicted alzRGs that are also DEGs according to human bulk RNA-seq studies with both late-stage AD (LAD) and control donors (STAR Methods).
(B) Violin plots show alzRGs are more likely differentially expressed in disease-associated microglia (DAM) according to mouse single-nucleus (GEO: GSE140511; three mice for each group; 5xFAD, wild-type, Trem2 knockout 5xFAD, and Trem2 knockout wild type) RNA-seq datasets (unpaired t test, t test statistic = 34.65, p = 2.59 × 10−206, numbers of replicates = 1,000).
(C) Violin plot shows alzRGs are more likely differentially expressed in disease-associated astrocyte (DAA) according to human prefrontal cortex single-nucleus (GEO: GSE157827; nine normal control and 12 AD human postmortem brain samples) RNA-seq dataset (unpaired t test, t test statistic = 52.01, p = 0.00, numbers of replicates = 1,000).
(D) Violin plot shows alzRGs are more likely differentially expressed in DAA according to human entorhinal cortex (EC) single-nucleus (GEO: GSE138852; six control and six AD human postmortem brain samples) RNA-seq dataset (unpaired t test, t test statistic = 15.18, p = 2.33 × 10−49, numbers of replicates = 1,000).
(E) Visualization of 32 predicted alzRGs that are also differentially expressed genes (DEGs) according to single-cell/nucleus RNA-seq studies collected from both mouse models and human postmortem brain tissues in two disease-associated immune subtypes, i.e., DAM and DAA (STAR Methods).