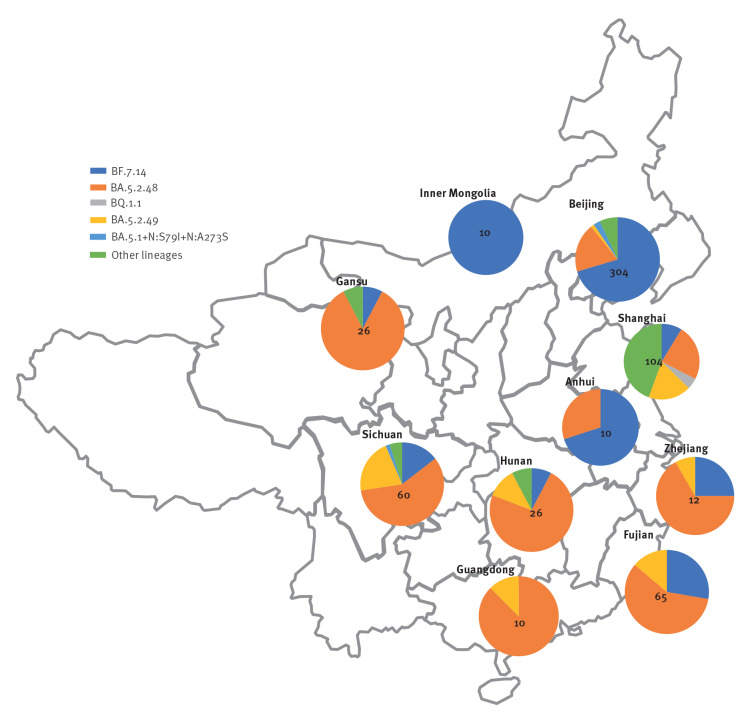
Figure 2.
Geographic distribution by province of Omicrona sub-lineages among sequencesb with case date after 1 September 2022 deposited from China in GISAID as of 11 January 2023 (n = 627 sequences)
a Omicron (Phylogenetic Assignment of Named Global Outbreak (Pango) lineage B.1.1.529).
b In the respective metadata, the deposited sequences were not labelled in as ‘imported’.
Other lineages include BA.5.2 (n = 9), BF.21 (n = 3), BF.7 (n =1), BA.5.1 (n = 8)in Beijing, BA.2 (n = 4), BA.5 (n = 1), BA.5.1 (n = 1), BA.5.2.1 (n = 4), other BA.5.2.* (n = 34), BE.1.4.2 (n = 1), BF.7 (n = 3), BQ.1 (n = 1), BQ.1.1 (n = 4), BQ.1.1.4 (n = 1), BQ.1.23 (n = 1), CH.1.1 (n = 1) in Shanghai, BA.5.2 in Gansu (n = 2), BA.5.2 in Huanan (n = 2), BA.5 (n = 1) and BA.5.2 (n = 1) in Guangdong, and BA.5.1 (n = 1) in Sichuan.
The disks (pie charts) are subdivided according to the proportion of each sub-lineage found. The numbers at the centres of the disks represent the total number of characterised viral sequences that originated from the provinces in question.