Fig. 1 ∣. ClampFISH 2.0 enables fast, cost-effective, exponential amplification of multiplexed RNA FISH signal in situ.
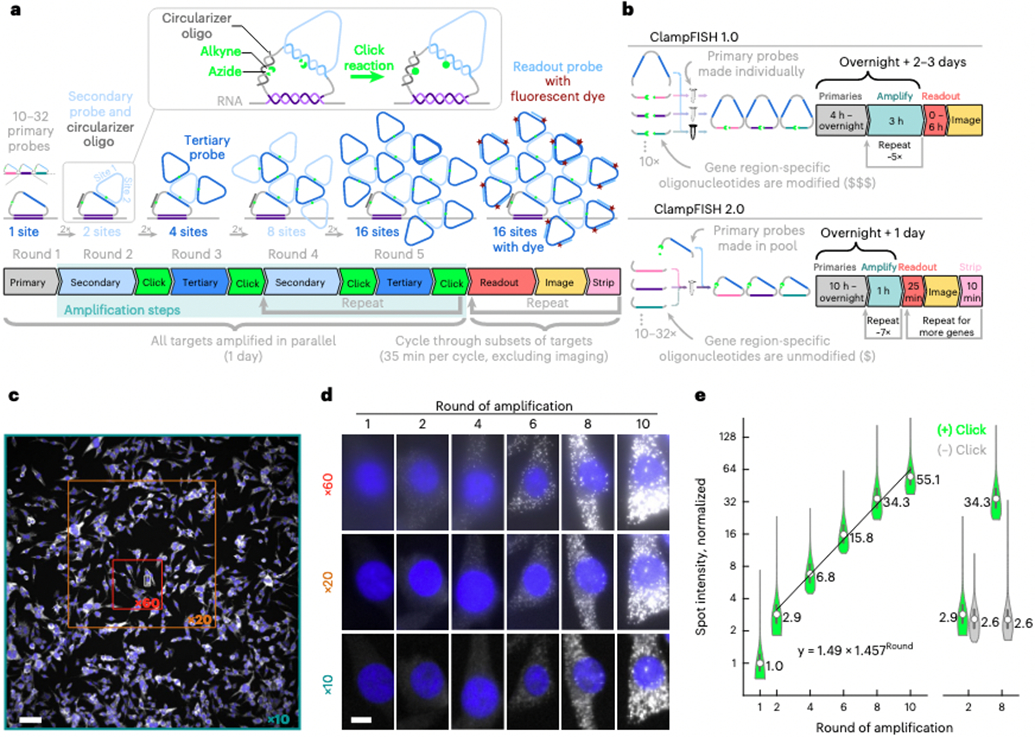
a, Schematic diagram of clampFISH 2.0. b, Comparison of clampFISH 2.0 and clampFISH 1.0: in clampFISH 2.0 the primary probes feature an inverted design, in which oligonucleotides modified for use with click chemistry can be re-used for all probes in any primary probe set. c, UBC clampFISH 2.0 at round 10 in WM989 A6-G3 cells, imaged with a ×10 objective, with the sizes of the smaller ×20 and ×60 fields of view overlaid. Scale bar, 100 μm. d, UBC clampFISH 2.0 in WM989 A-G3 cells shown at progressively higher rounds of amplification at ×60, ×20 and ×10 magnification. The experiment was performed twice with similar results. Scale bar, 10 μm. e, Left: UBC clampFISH 2.0 spot intensity (normalized to the median intensity from round 1) over progressively higher rounds of amplification, with the normalized median intensity from rounds 2, 4, 6, 8 and 10 fitted to an exponential curve (n = 70,800, 64,440, 47,520, 49,800, 53,760 and 53.280 spots from each of rounds 1, 2, 4, 6, 8 and 10, respectively, from the same experiment). Right: spot intensity at rounds 2 and 8 when the copper catalyst is included (green) or not included (gray) in the click reaction (n = 59,520 and 57,120 spots in rounds 2 and 8, respectively, when the copper catalyst was not included, from the same experiment). The circles represent the median, and the bounds of the inner box plots represent the 25th and 75th percentiles. The experiment was performed twice. See Extended Data Fig. 4 and Supplementary Fig. 3 for data associated with all targets and amplifier sets from both replicates.
