Extended Data Fig. 9∣. clampFISH 2.0 scaffolds remain stably bound after multiple rounds of readout stripping and storage at 4 °C for 4 months.
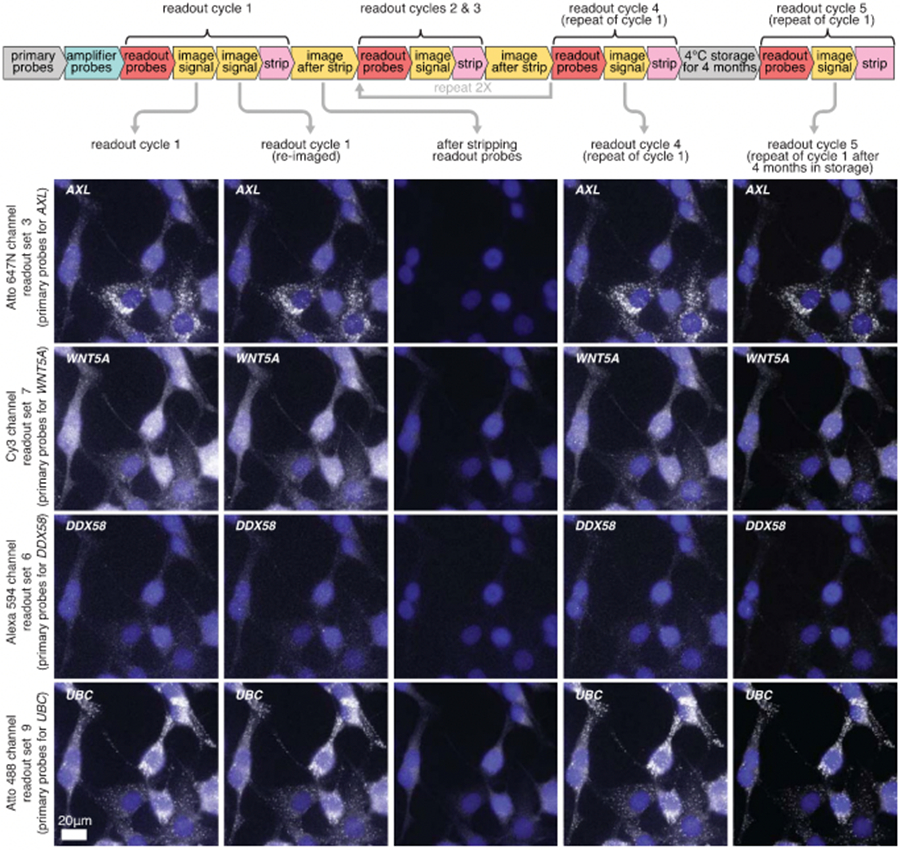
Images of clampFISH 2.0 spots from a 20X objective over readout cycles where we repeatedly use 4 sets of readout probes which label (from top to bottom) AXL, WNTSA, DDX58, and UBC clampFISH 2.0 scaffolds. Column 1: readout cycle 1. Column 2: readout cycle 1, re-imaged after removing the sample from the microscope stage and stored overnight at 4 °C. Column 3: after stripping off readout probes from readout cycle 1. Column 4: readout cycle 4, where we repeat readout cycle 1 after readout cycles 2 and 3 (where different sets of genes were labeled). Column 5: readout cycle 5, performed after storing the sample at 4 °C in 2X SSC for 4 months. DAPI overlay is contrasted separately for each column. Each row of readout cycle S (column 5) is contrasted with 180% the intensity range of the first four columns. The cycle S signal presumably appeared brighter due to changes in the microscope’s optics during that time frame (for example, greater sample illumination or increased transmission to the sensor). The experiment was performed twice with similar results, except for column 5 data (after 4 months storage) which was performed once. See Supplementary Figs. 14 and 15 for quantification of each experimental replicate.
