Extended Data Fig. 1∣. clampFISH 2.0 amplifies GFP RNA FISH signal with high specificity.
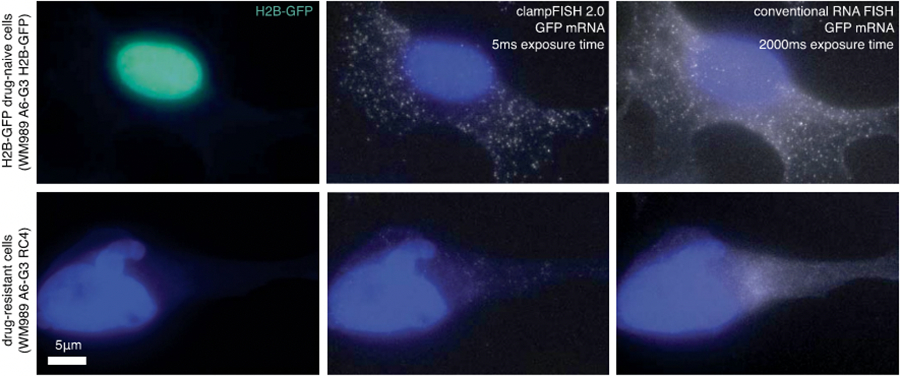
Images of GFP clampFISH 2.0 spots in drug-naive H2B GFP VVM989 A6-G3 cells (top) and vemurafenib-resistant WM989 A6-G3 RC4 cells (bottom) with a 20 nucleotide secondary-targeting readout probe (labeled with Atto 647N) and conventional single-molecule RNA FISH probes (labeled with Alexa 555) targeting different regions of the same RNA. Shown are maximum intensity projections of 9 z-planes at 60X magnification. Shown are data using amplifier set 1, where 15 amplifier sets were tested in total. The experiment was performed once. See Supplementary Fig. 2 for co-localization quantification of all amplifier sets. For details, see Supplementary Methods description of amplifier screen experiment. As expected, we observed bright GFP clampFISH 2.0 spot counts in cells with nuclear-localized GFP signal, but not in cells without the H2B-GFP construct.
