Extended Data Fig. 2∣. clampFISH 2.0 amplifies EGFR RNA FISH signal with high accuracy.
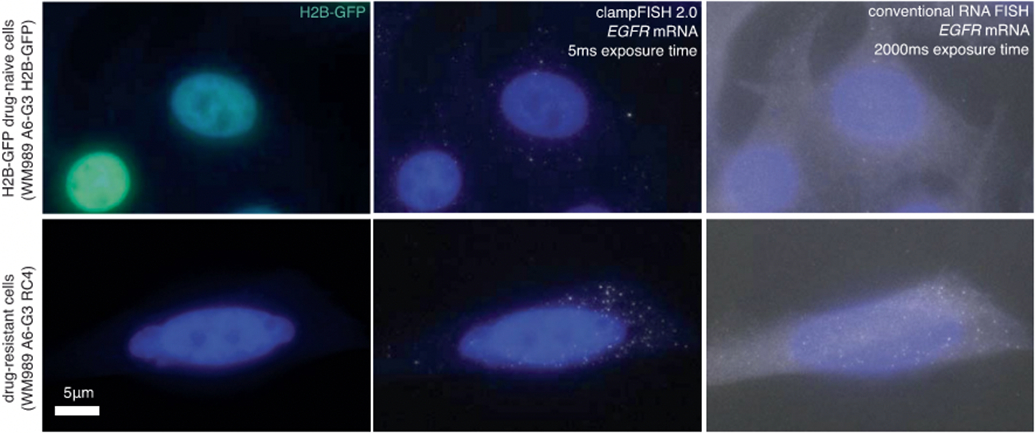
Images of EGFR clampFISH 2.0 spots in drug-naive H2B-GFP WM989 A6-G3 cells (top) and vemurafenib-resistant WM989 A6-G3 RC4 cells (bottom) with a 20 nucleotide secondary-targeting readout probe (labeled with Atto 647N) and conventional single-molecule RNA FISH probes (labeled with Cy3) targeting different regions of the same RNA. Shown are maximum intensity projections of 9 z-planes at 60X magnification. Shown are data using amplifier set 1, where 15 amplifier sets were tested in total. The experiment was performed once. For details, see Supplementary Methods description of amplifier screen experiment. As expected from bulk RNA sequencing data, we observed many more EGFR clampFISH 2.0 spots in the vemurafenib-resistant cells than in the drug-naive cells.
