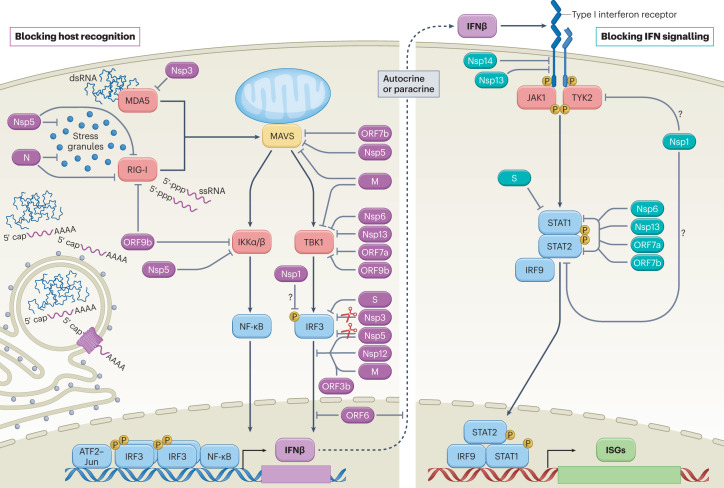
Fig. 2. SARS-CoV-2-mediated interference of cellular innate immune signalling.
Virus infection generates replication intermediates and/or induces the formation of stress granules that serve as platforms for RIG-I-like receptor (RIG-I or MDA5) activation. Host recognition of viral pathogen-associated molecular patterns, such as single-stranded RNA (ssRNA) with an exposed 5′-triphosphate or double-stranded RNA (dsRNA), promotes the assembly of a mitochondria-localized signalling hub orchestrated by mitochondrial antiviral signalling protein (MAVS), and culminates in the activation of host kinases IKKα, IKKβ and TBK1. Kinase activation induces the production of interferon-β (IFNβ) through cooperative engagement of the ATF2–JUN, interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB) transcription factors. Secreted IFNβ functions in an autocrine or paracrine manner to promote an antiviral state in cells. On binding of IFNβ, the type I IFN receptor subunits on the cell surface dimerize, bringing together the receptor-associated kinases, Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which subsequently activate each other via transphosphorylation and promote the recruitment and phosphorylation of the signal transducer and activator of transcription (STAT) molecules, STAT1 and STAT2. Phosphorylated STAT1 and STAT2 form a stable complex with interferon regulatory factor 9 (IRF9) that translocates into the nucleus, where it promotes the transcription of IFN-stimulated genes (ISGs). Each of these processes is the target of SARS-CoV-2 interference, as illustrated here and further described in Table 1. Viral proteins that inhibit aspects of host recognition and the associated signalling pathways are shown in purple, whereas those that block components of the IFN signalling pathway are depicted in teal. Owing to the ability of Nsp1 to more generally inhibit protein synthesis, its role in specifically blocking elements of these pathways remains uncertain (question marks). M, membrane protein; N, nucleocapsid protein; P, phosphorylation; S, spike protein.