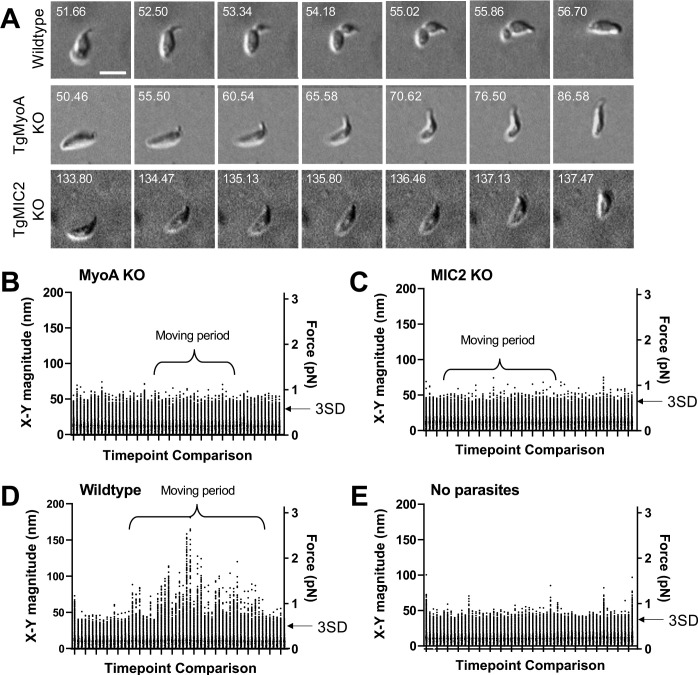
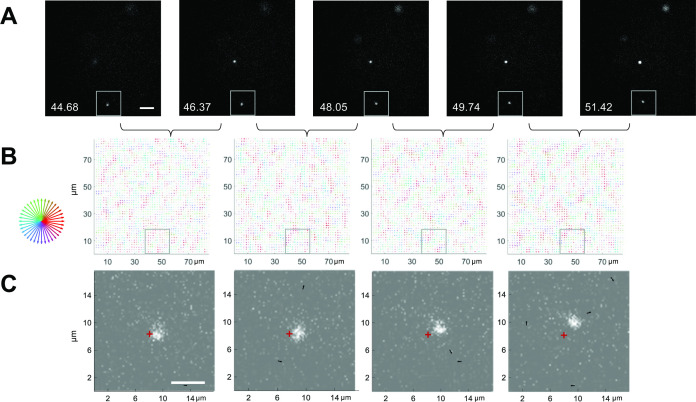
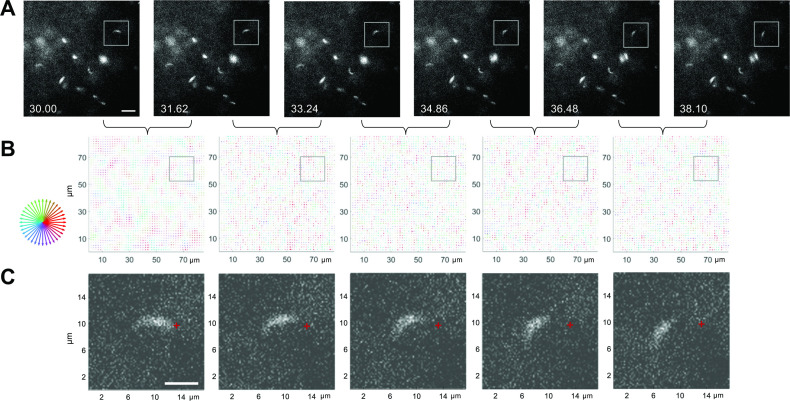
Figure 6. 3D motility and force mapping of parasites lacking TgMyoA or TgMIC2.
(A) Brightfield images showing TgMyoA knockout and TgMIC2 knockout parasites moving within a Matrigel matrix without a detectable constriction. A wild-type parasite undergoing a typical constriction is shown for comparison. The TgMyoA knockout parasite makes right-angled ‘stairstep’ turn, and the TgMIC2 knockout parasite moves erratically in a tightly twisting arc. Scale bar = 5 µm, timestamps in seconds. See Videos 11 and 12 for the entire time series for the TgMyoA and TgMIC2 knockout parasites. (B–E) As in Figure 3A, the plots show the magnitudes of the 16,807 fibrin x-y displacement vectors between each pair of successive image volumes generated in experiments using: (B) TgMyoA knockout parasites; (C) TgMIC2 knockout parasites; (D) wild-type parasites; and (E) no added parasites (fibrin only). Periods during the 96-second time course when parasites were moving are indicated. For each time course, the two consecutive time points that gave the lowest mean displacement magnitude were used to set the background threshold: any displacements less than three standard deviations (3SD) above this mean were considered noise for that dataset. Examples of the force maps surrounding motile TgMyoA and TgMIC2 knockout parasites are shown in Figure 6—figure supplements 1 and 2, respectively.