Figure 1. Community-science-designed RNA datasets from the Eterna “Cloud Lab” experiments identify consistent discrepancies in ensemble calculations from secondary-structure packages.
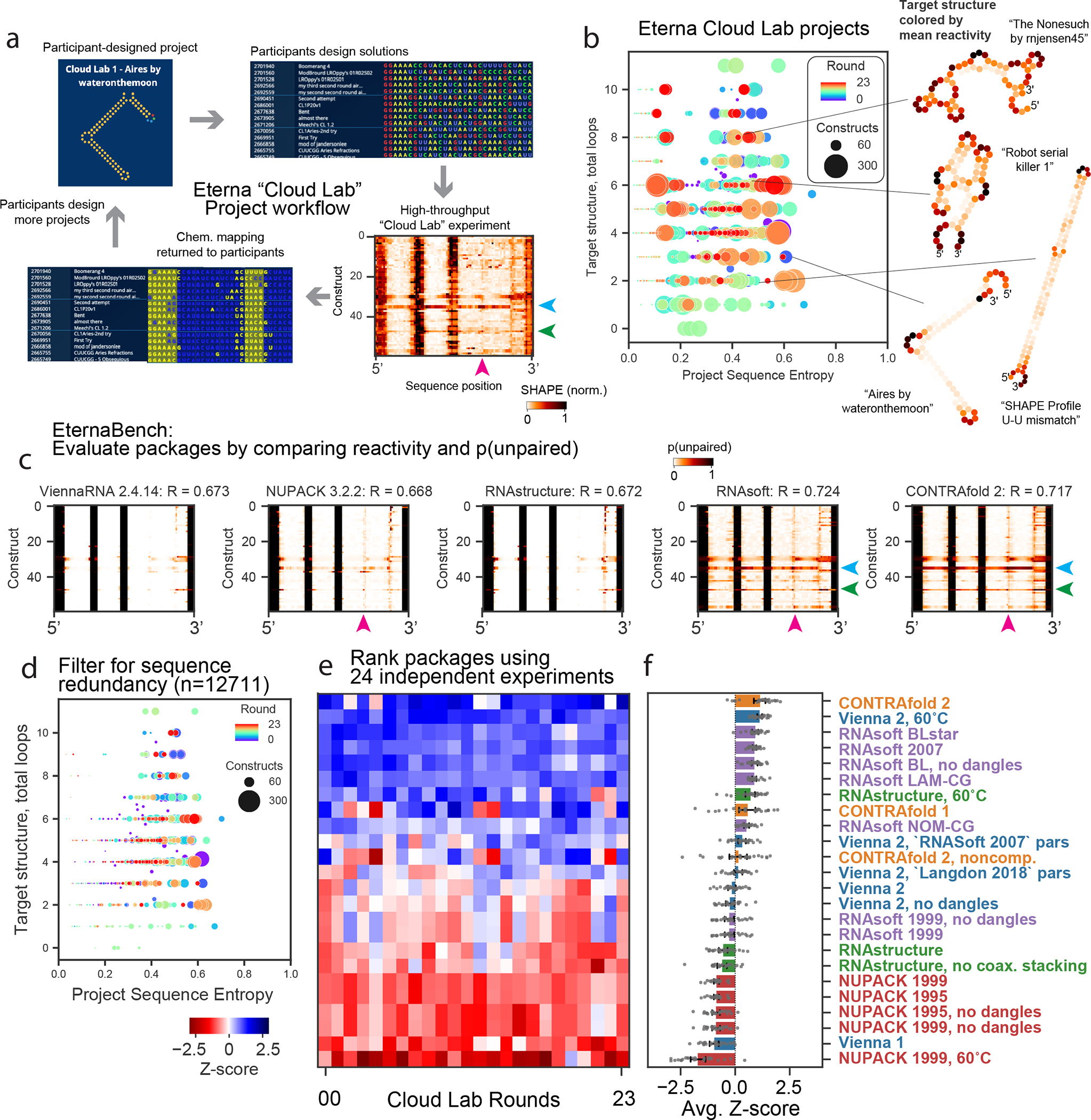
(A) Workflow of cloud lab rounds: Eterna participants design “projects”, typically intended as RNA design challenges. Players submit solutions, all of which are synthesized in high-throughput via MAP-seq experiments. Example reactivity data are depicted from the project “Aires” by participant wateronthemoon. Data are returned to participants in the in-game browser, which served as the basis for more player-designed projects.
(B) Calculating the average positional entropy for all solutions collected for each project reveals that participants were able to design a diverse set of solutions, independent of target structure complexity (monitored as number of loops in the target structure). Example target structures are colored by average reactivity. (C) Example unpaired probabilities for 60 example constructs from the project “Aires”, for which reactivity data are shown in (A), across 5 representative secondary structure packages. Blue, green, magenta arrows indicate package predictions that recapitulate experimental partially reactive features. CONTRAfold and RNAsoft predictions for p(unpaired) have higher correlation to experimental reactivity data. (D) Analogous representation to (B) for the redundancy-filtered EternaBench dataset. (E) We compared many commonly used packages and secondary structure prediction options over 24 Cloud Lab independent experiments. We calculated the Pearson correlation coefficient and calculated the Z-score across all packages evaluated for each dataset. (F) Final ranking is obtained by averaging the Z-scores obtained across all datasets. Error bars represent 95% confidence interval of the mean obtained over 1000 iterations of bootstrapping over n= 24 independent experiments, which comprised 12,711 independent constructs total.
