Extended Data Fig. 9. Extended data corresponding to EternaFold development and test set evaluation.
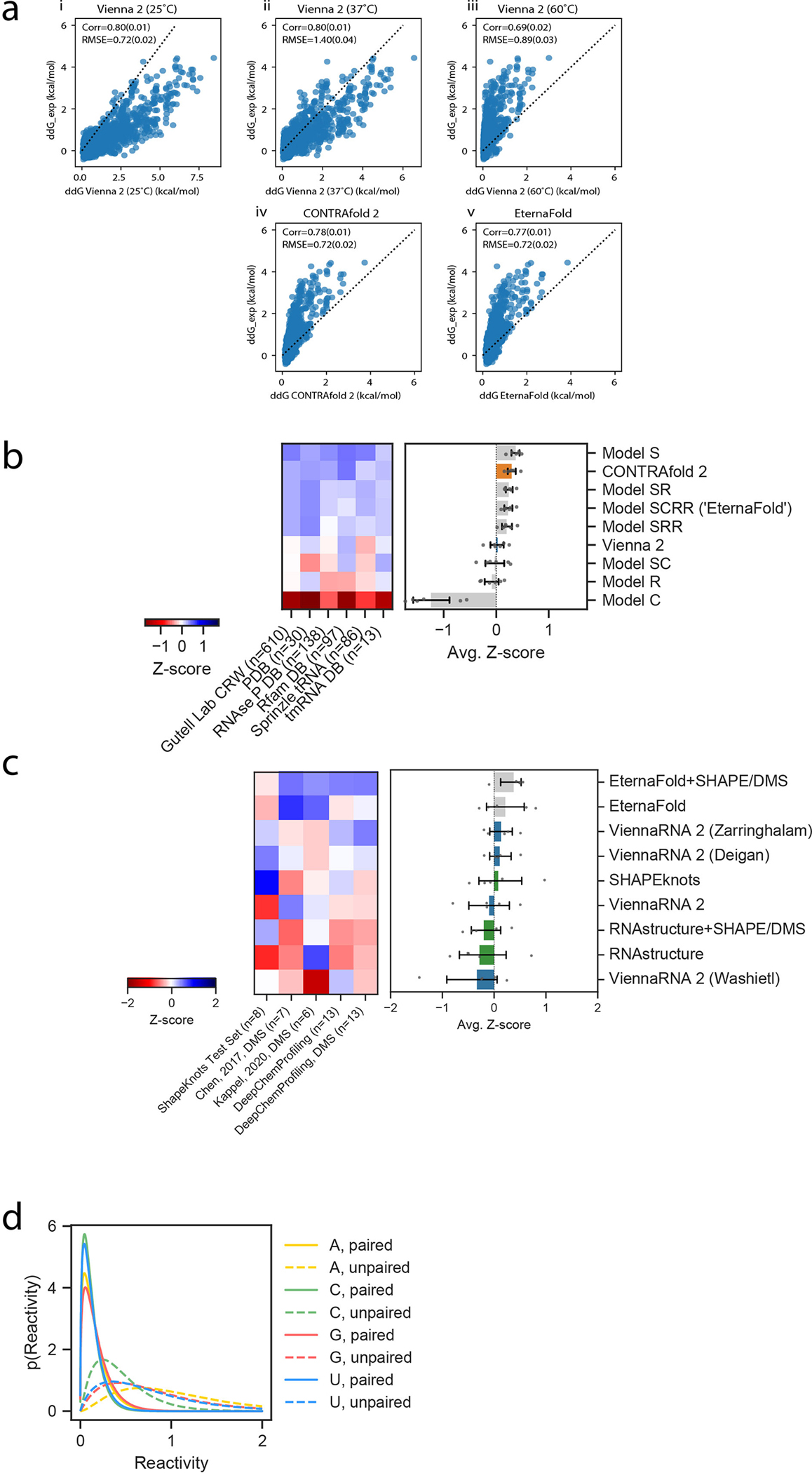
a) Comparing Vienna, CONTRAfold, and EternaFold predictions in predicting free energy of PUM binding. i) Replication of ddG_exp for both PUM WT and mutant binding from (Becker, 2019). The same calculation in Vienna 2 at 37°C shows lower Root-mean-squared error (RMSE) (ii), but higher RMSE at 60°C (iii). CONTRAfold 2 shows no improvement over Vienna at 37°C (iv), but EternaFold shows modest improvement over both (v). b) Package performance for the S-Processed test set is qualitatively similar to results on the ArchiveII-NR test set (cf. Fig. 3b). Error bars represent 95% confidence interval of the mean calculated with 1000 iterations of bootstrapping over n=6 independent datasets, which contain 974 independent constructs total. C) Evaluating SHAPE- and DMS- directed folding. Error bars represent 95% confidence interval of the mean calculated with 1000 iterations of bootstrapping over n=5 independent datasets of RNAs with known secondary structures,, which contain 47 constructs total. d) Potentials learned from EternaFold training and used in SHAPE-directed structure prediction.
