Figure 4. EternaFold improved prediction extends across diverse natural RNA contexts and experiments.
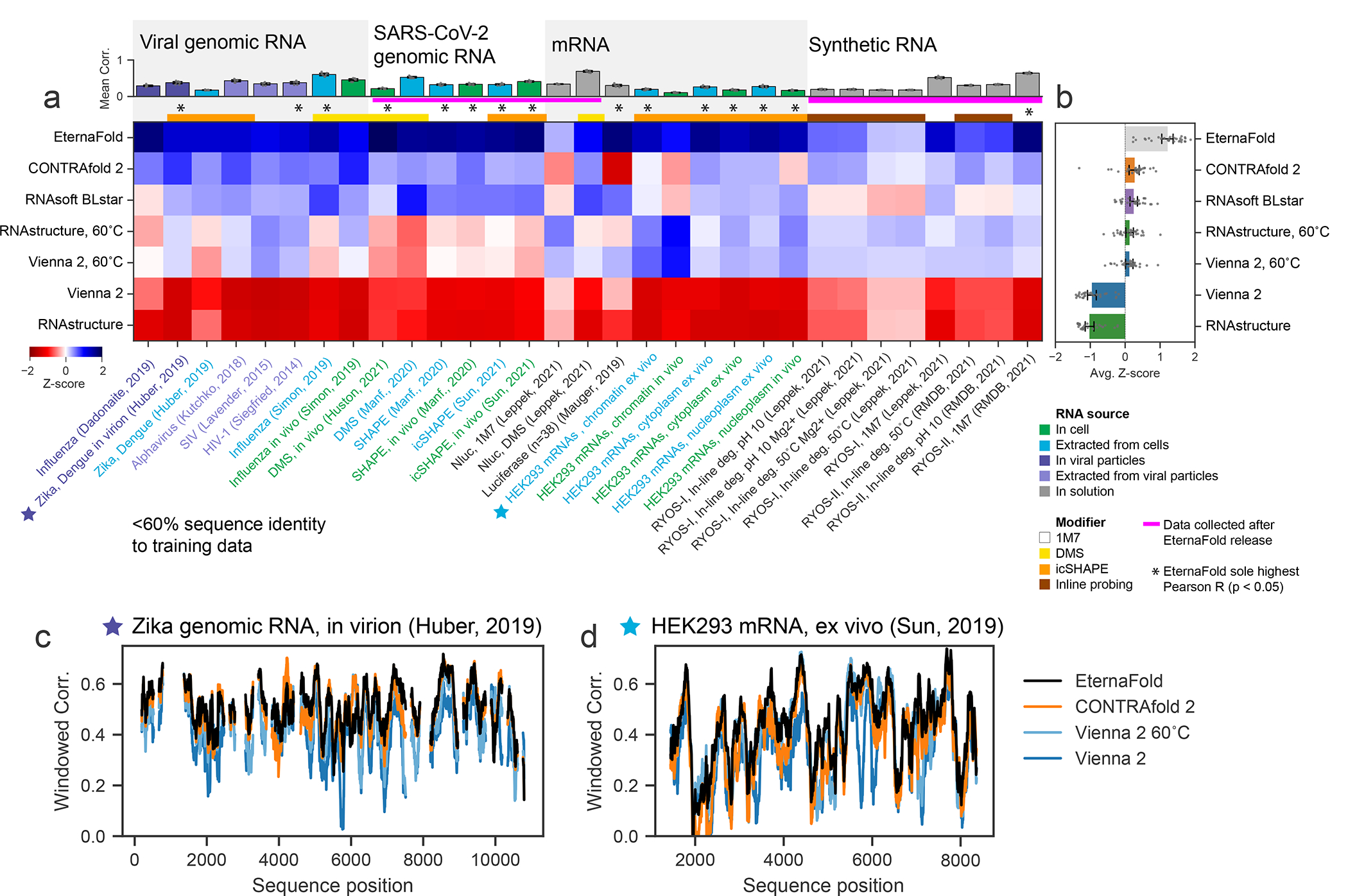
A) Mean package correlation for top-performing representative packages selected to benchmark for 31 independent chemical mapping datasets from a variety of biological contexts and with other chemical modifiers. Each dataset was filtered to contain sequences with <60% sequence similarity to the EternaFold training set. (B) EternaFold is ranked highest in average Z-score. Error bars represent 95% confidence interval of the mean obtained over 1000 iterations of bootstrapping over n=31 datasets from literature. Calculating correlations over sequence windows indicates that EternaFold demonstrates uniformly higher correlation across sequence position for two representative datasets: (C) Zika genome probed in virion (ref. 45), (D) Human mRNA ENST00000495843 probed ex vivo (ref. 53).
