Extended Data Fig. 1. Extended analysis of package rankings based on Eterna Cloud lab chemical mapping data.
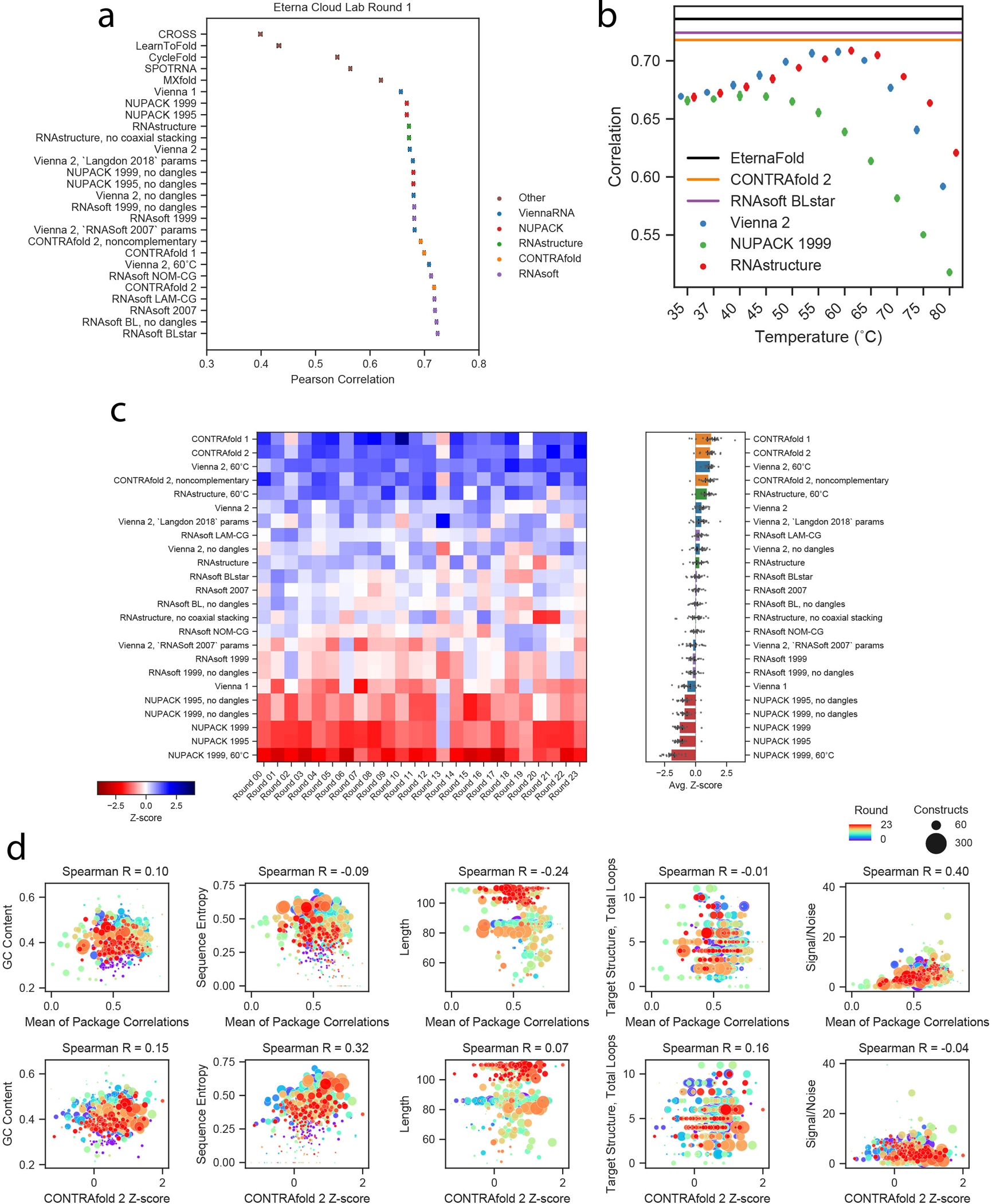
a) Pearson correlation of all package options tested on Cloud Lab Round 1, which was also a holdout test set for EternaFold training studies. Mean ± SEM of Pearson correlation calculated via bootstrapping, n=1088 independent constructs. b) ViennaRNA 2, NUPACK 1999, and RNAstructure show maximum Pearson correlation to chemical mapping data at 60°C, 40°C, and 60°C respectively for Eterna Cloud Lab Round 1. Mean ± SEM of Pearson correlation calculated via bootstrapping, n=1088 independent constructs. c) Ranking across Cloud lab dataset rounds using Spearman rank correlation (compare to Figure 1E,F). Error bars represent 95% confidence interval of the mean obtained over 1000 iterations of bootstrapping over 24 independent experiments, n=12,711 independent constructs total. d) (Top) Mean Pearson correlations, calculated over each project (as opposed to each dataset), compared to sequence metrics of the Cloud Lab projects. The strongest correlation to mean correlation was Signal/Noise ratio. (Bottom) Z-score of CONTRAfold-2, calculated over each project, compared to sequence metrics of the Cloud Lab projects.
